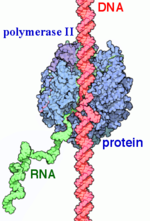
User:Neal Hayhurst/RNA Polymerase II/Sandbox 1
From Proteopedia
< User:Neal Hayhurst(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 72: | Line 72: | ||
====Reinitiation==== | ====Reinitiation==== | ||
| - | Some of the TFs still remain on the DNA to reinitiate transcription and to mark where transcription has just occurred<ref name="F">Hahn, S. Structure and Mechanism of the RNA Polymerase II Transcription Machinery. Nature Structure and Molecular Biology. 2004, 11, 394-403.</ref>. These remaining transcription factors are collectively called the <scene name='86/862212/ | + | Some of the TFs still remain on the DNA to reinitiate transcription and to mark where transcription has just occurred<ref name="F">Hahn, S. Structure and Mechanism of the RNA Polymerase II Transcription Machinery. Nature Structure and Molecular Biology. 2004, 11, 394-403.</ref>. These remaining transcription factors are collectively called the <scene name='86/862212/The_real_scaffold_1/3'>scaffold complex</scene>. The scaffold complex consists of TFIIH, TFIIE, TFIID, and TFIIA and helps bypass the slow step of recruiting these TFs for reinitiation<ref name="F">Hahn, S. Structure and Mechanism of the RNA Polymerase II Transcription Machinery. Nature Structure and Molecular Biology. 2004, 11, 394-403.</ref>. Thus, the scaffold complex only needs to recruit TFIIF, TFIIB, and RNAP II to restart transcription to synthesize another RNA product<ref name="F">Hahn, S. Structure and Mechanism of the RNA Polymerase II Transcription Machinery. Nature Structure and Molecular Biology. 2004, 11, 394-403.</ref>. |
===Elongation=== | ===Elongation=== | ||
| Line 83: | Line 83: | ||
====Kinetics==== | ====Kinetics==== | ||
| - | Multiple kinetic models have been proposed with largely a consensus on some form of a Brownian ratchet mechanism involving the trigger loop and bridge helix in which the RNAPII has both forward and backward motion with a preference for forward translocation<ref>DOI: 10.1016/j.cell.2004.11.045</ref><ref name="Mishanina" />. | + | Multiple kinetic models have been proposed with largely a consensus on some form of a Brownian ratchet mechanism involving the trigger loop and bridge helix in which the RNAPII has both forward and backward motion with a preference for forward translocation<ref>DOI: 10.1016/j.cell.2004.11.045</ref><ref name="Mishanina" />. The net forward rate of RNAP II is about 2kb/min in vivo. RNAPII has nearly infinite processivity due to the Rpb2 <scene name='86/862212/Clamp/2'>clamp</scene> subunit swinging down over DNA to trap it in the cleft<ref name="VVP" />. |
| - | + | ||
| - | RNAPII has nearly infinite processivity due to the Rpb2 <scene name='86/862212/Clamp/2'>clamp</scene> subunit swinging down over DNA to trap it in the cleft<ref name="VVP" />. | + | |
Current revision
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Young RA. RNA polymerase II. Annu Rev Biochem. 1991;60:689-715. doi: 10.1146/annurev.bi.60.070191.003353. PMID:1883205 doi:http://dx.doi.org/10.1146/annurev.bi.60.070191.003353
- ↑ 2.0 2.1 Myer VE, Young RA. RNA polymerase II holoenzymes and subcomplexes. J Biol Chem. 1998 Oct 23;273(43):27757-60. doi: 10.1074/jbc.273.43.27757. PMID:9774381 doi:http://dx.doi.org/10.1074/jbc.273.43.27757
- ↑ 3.0 3.1 3.2 Sobennikova MV, Shematorova EK, Shpakovskii GV. [C-terminal domain (CTD) of the subunit Rpb1 of nuclear RNA polymerase II and its role in the transcription cycle]. Mol Biol (Mosk). 2007 May-Jun;41(3):433-49. PMID:17685222
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 RNA polymerase II transcription initiation: A structural view D. B. Nikolov, S. K. Burley Proceedings of the National Academy of Sciences Jan 1997, 94 (1) 15-22; DOI: 10.1073/pnas.94.1.15
- ↑ Hurwitz J. The discovery of RNA polymerase. J Biol Chem. 2005 Dec 30;280(52):42477-85. doi: 10.1074/jbc.X500006200. Epub 2005, Oct 17. PMID:16230341 doi:http://dx.doi.org/10.1074/jbc.X500006200
- ↑ doi: https://dx.doi.org/10.1038/nrm1796
- ↑ 7.0 7.1 7.2 7.3 Orphanides, George, Thierry Lagrange, and Danny Reinberg. The general transcription factors of RNA polymerase II. Genes & development 10.21. 1996. 2657-2683
- ↑ 8.0 8.1 Xin, L.; Bushnell, D. A.; and Kornburg, R. D. RNA Polymerase II Transcription: Structure and Mechanism. Biochemica et Biophysica Acta. 2013, 1829, 2-8.
- ↑ Chang-Hui Shen; Diagnostic Molecular Biology, 2019
- ↑ He, Yuan, et al. Near-atomic resolution visualization of human transcription promoter opening. Nature 533.7603. 2016.
- ↑ 11.0 11.1 11.2 11.3 11.4 11.5 11.6 11.7 11.8 Hahn, S. Structure and Mechanism of the RNA Polymerase II Transcription Machinery. Nature Structure and Molecular Biology. 2004, 11, 394-403.
- ↑ 12.0 12.1 Eick, D, Geyer, M.The RNA Polymerase II Carboxy-Terminal Domain (CTD) Code. Chemical Reviews, 2013, 113 (11), 8456-8490 DOI: 10.1021/cr400071f
- ↑ Wang W, Carey M, Gralla JD. Polymerase II Promoter Activation: Closed Complex Formation and ATP-Driven Start Site Opening. Science. 1992;255:450–453.
- ↑ 14.0 14.1 14.2 14.3 14.4 14.5 14.6 14.7 14.8 14.9 Voet, D., Voet, J. G., & Pratt, C. W. (2013). Transcription and RNA Processing. In Fundamentals of biochemistry: life at the molecular level (pp. 933–942). Wiley.
- ↑ Nudler, E. RNA Polymerase Active Center: The Molecular Engine of Transcription. Annu. Rev. Biochem. 2009, 78, 335-361.
- ↑ Svetlov, V., & Nudler, E. (2013). Basic mechanism of transcription by RNA polymerase II. Biochimica et biophysica acta, 1829(1), 20–28. https://doi.org/10.1016/j.bbagrm.2012.08.009
- ↑ Castano E, Gross P, Wang Z, Roeder RG, Oelgeschlager T. The C-terminal domain-phosphorylated IIO form of RNA polymerase II is associated with the transcription repressor NC2 (Dr1/DRAP1) and is required for transcription activation in human nuclear extracts. Proc Natl Acad Sci U S A. 2000 Jun 20;97(13):7184-9. doi: 10.1073/pnas.140202297. PMID:10852970 doi:http://dx.doi.org/10.1073/pnas.140202297
- ↑ doi: https://dx.doi.org/https
- ↑ 19.0 19.1 19.2 19.3 19.4 19.5 19.6 19.7 19.8 Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006 Dec 1;127(5):941-54. PMID:17129781 doi:http://dx.doi.org/10.1016/j.cell.2006.11.023
- ↑ 20.0 20.1 20.2 20.3 20.4 Mishanina TV, Palo MZ, Nayak D, Mooney RA, Landick R. Trigger loop of RNA polymerase is a positional, not acid-base, catalyst for both transcription and proofreading. Proc Natl Acad Sci U S A. 2017 Jun 27;114(26):E5103-E5112. doi:, 10.1073/pnas.1702383114. Epub 2017 Jun 12. PMID:28607053 doi:http://dx.doi.org/10.1073/pnas.1702383114
- ↑ 21.0 21.1 Da LT, Wang D, Huang X. Dynamics of pyrophosphate ion release and its coupled trigger loop motion from closed to open state in RNA polymerase II. J Am Chem Soc. 2012 Feb 1;134(4):2399-406. doi: 10.1021/ja210656k. Epub 2012 Jan , 24. PMID:22206270 doi:http://dx.doi.org/10.1021/ja210656k
- ↑ Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005 Jan 28;120(2):183-93. doi: 10.1016/j.cell.2004.11.045. PMID:15680325 doi:http://dx.doi.org/10.1016/j.cell.2004.11.045
- ↑ 23.0 23.1 23.2 23.3 Schreieck A, Easter AD, Etzold S, Wiederhold K, Lidschreiber M, Cramer P, Passmore LA. RNA polymerase II termination involves C-terminal-domain tyrosine dephosphorylation by CPF subunit Glc7. Nat Struct Mol Biol. 2014 Feb;21(2):175-179. doi: 10.1038/nsmb.2753. Epub 2014, Jan 12. PMID:24413056 doi:http://dx.doi.org/10.1038/nsmb.2753
- ↑ doi/10.1101/gad.1055503
- ↑ Shah N, Maqbool MA, Yahia Y, El Aabidine AZ, Esnault C, Forne I, Decker TM, Martin D, Schuller R, Krebs S, Blum H, Imhof A, Eick D, Andrau JC. Tyrosine-1 of RNA Polymerase II CTD Controls Global Termination of Gene Transcription in Mammals. Mol Cell. 2018 Jan 4;69(1):48-61.e6. doi: 10.1016/j.molcel.2017.12.009. PMID:29304333 doi:http://dx.doi.org/10.1016/j.molcel.2017.12.009
- ↑ 26.0 26.1 26.2 26.3 26.4 26.5 doi/10.1073/pnas.251664698
- ↑ 27.0 27.1 27.2 Brueckner F, Cramer P. Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation. Nat Struct Mol Biol. 2008 Aug;15(8):811-8. Epub 2008 Jun 13. PMID:18552824 doi:10.1038/nsmb.1458
Alpha-aminitin chemical structure image courtesy of https://en.wikipedia.org/wiki/Alpha-Amanitin#/media/File:Alpha-amanitin_structure.png
Notes
From structural components:
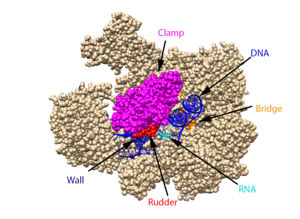
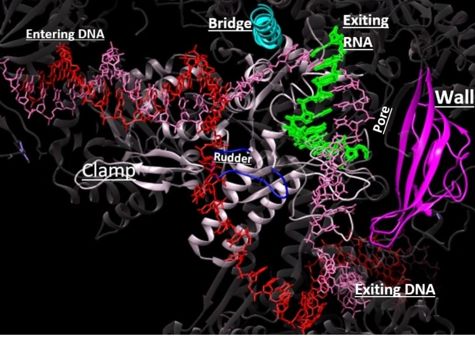
Structural overview: [PDB: 5VVR: with highlighted sections mentioned below]
Bridge: Depicted: [PDB: 1I6H: 810-845.a]
Wall: Depicted: [PDB: 1R5U: 853-919.b; 933-972.b]
Clamp: Depicted: [PDB: 1R5U: 3-345.a; 1395-1435.a; 1158-1124.b]
Rudder: Depicted: [PDB: 5VVR: 306-321.a]
Content Donators
This page was created as a final project for the Advanced Biochemistry course at Wabash College during the Fall of 2019 and Fall of 2020. This page was reviewed by Dr. Wally Novak of Wabash College.