Sandbox Reserved 1639
From Proteopedia
(Difference between revisions)
| (29 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
{{Sandbox_Reserved_BHall_F20}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | {{Sandbox_Reserved_BHall_F20}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
| - | == | + | ==Arabidopsis thaliana legumain isoform beta in zymogen state (6YSA)== |
<StructureSection load='6YSA' size='340' side='right' caption='Caption for this structure' scene=''> | <StructureSection load='6YSA' size='340' side='right' caption='Caption for this structure' scene=''> | ||
This is a default text for your page ''''''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | This is a default text for your page ''''''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
| Line 6: | Line 6: | ||
== Function of your Protein == | == Function of your Protein == | ||
| + | |||
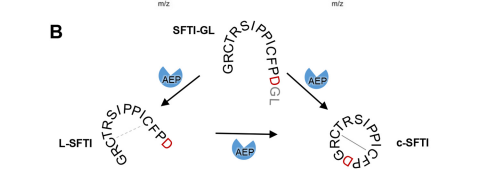
| + | My protein, legumain isoform beta (proAtLEGb), plays a very important role in the plant ''Arabidopsis thaliana.'' The specific function of the protein is important for the processing and maturation of the seed storage proteins within storage vacuoles and the programmed cell death and may functionally substitute for the caspases (not found in plants). Our protein is an enzyme and known substrates for it include the pro12Sglobulin and pro2S albumin proteins. The formation of the product is formed from cleavage at the active site by a special activation peptide (AP). The product then takes a cyclic formation after cleavage. More about the active site will be discussed under the amino acid section. | ||
== Biological relevance and broader implications == | == Biological relevance and broader implications == | ||
| + | This protein is important for this organism because it deals with seed maturation and how the plant eventually dies. | ||
| + | There really aren't any abnormal events associated with the protein that is known. | ||
| + | Our protein is the first example of a plant legumain capable of linking free termini, which is important because the discovery of these isoform-specific differences will allow us to identify and rationally design efficient ligases with application in biotechnology and drug development. | ||
== Important amino acids == | == Important amino acids == | ||
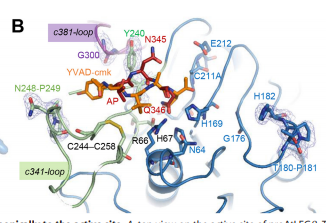
| - | + | To dig deeper and better understand the protein, you can see where the <scene name='86/861621/Protein_view_2/3'>ligand is bound to the protein</scene>. However, these ligands aren't entirely that important to the active site. The ligands shown are there to help stabilize the structure. The actual important enzyme of the active site is actually the activation peptide(AP). This AP [[Image:AP.PNG]] is made up of amino acid residues of ASN345 (asparagine), GLN346 (glutamine), ARG347 (arginine), which form<scene name='86/861621/Catalytic_triad/1'> the catalytic triad</scene> and where the cleavage happens. This cleavage [[Image:Cleavage.png]] basically splits the sequence in half of prime and nonprime on either side of the split. Then, the split sequences then take on a cyclic formation. This cyclic formation is crucial for the protein to remain in homeostasis and is the main source of energy transformation with bonds being broken and recreated during cyclization. | |
| + | Our protein is also very <scene name='86/861621/Hydrophobic_parts/1'>hydrophobic at certain ends.</scene> [[Image:Hydrophobic.png]] | ||
== Structural highlights == | == Structural highlights == | ||
| + | |||
| + | Looking at the <scene name='86/861621/Secondary_structure/1'>secondary structure</scene>, there are 10 alpha-helices, a parallel beta-sheet, 2 anti-parallel beta-sheets, and about 7 random coils. | ||
| + | The parallel beta-sheet is where the interaction with the ligand (citric acid) happens. The His138 amino acid residue forms an interaction at this parallel beta-sheet. | ||
| + | For the <scene name='86/861621/Protein_view_2/3'>tertiary structure</scene>, there is 1 polymer with plenty of hydrogen bonds, disulfide bridges, and hydrophobic interactions. It also contains sulfate ions, a citric acid, and a carbohydrate (glucopyranose). | ||
| + | The <scene name='86/861621/Quartenary_structure/1'>quaternary structure</scene> you have already seen, our protein consists of 12 total chains. | ||
== Other important features == | == Other important features == | ||
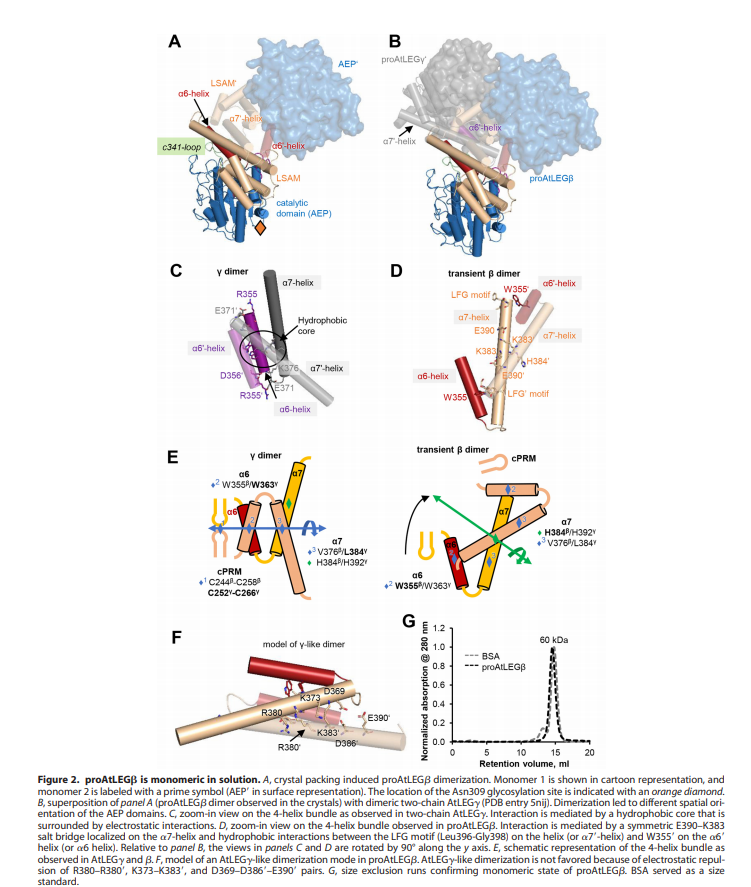
| + | An important feature is that proAtLEGb forms atypical dimers in the crustal and is monomeric in solutions. When compared to another form of our protein AtLEGgy findings suggest that the observed beta-dimer is weak and probably only transient in solution. [[Image:Monomeric.png]] | ||
| - | + | Another important feature is the cyclization of the peptides after cleavage. The cyclization is important for the cell stays programmed and is the same every time. [[Image:Cyclization.png]] | |
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
| - | <references/> | + | <references/> |
| + | <ref>32719006</ref> | ||
| + | PMID:32719006 | ||
Current revision
| This Sandbox is Reserved from 09/18/2020 through 03/20/2021 for use in CHEM 351 Biochemistry taught by Bonnie Hall at Grand View University, Des Moines, IA. This reservation includes Sandbox Reserved 1628 through Sandbox Reserved 1642. |
To get started:
More help: Help:Editing |
Arabidopsis thaliana legumain isoform beta in zymogen state (6YSA)
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
[1] PMID:32719006