Sandbox Reserved 1656
From Proteopedia
(Difference between revisions)
(New page: {{Sandbox_Reserved_ESBS20_}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> ==Your Heading Here (maybe something like 'Structure')== <StructureSection load='1stp' size='340' side='right' capti...) |
|||
| (98 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
{{Sandbox_Reserved_ESBS20_}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | {{Sandbox_Reserved_ESBS20_}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
| - | == | + | ==Deubiquitinases== |
| - | <StructureSection load=' | + | <StructureSection load='3TMP' size='340' side='right' caption='The catalytic domain of human deubiquitinase DUBA in complex with ubiquitin aldehyde' scene=''> |
| - | + | '''Deubiquitinating enzymes or Deubiquitinases (DUBs)''' are [https://en.wikipedia.org/wiki/Enzyme enzymes] with an ubiquitin-dependent action. More than a hundred DUBs genes are known to be encoded by the humans genome, making it a very large [https://en.wikipedia.org/wiki/Protein protein] family. This diversity allows targeted actions and a certains specificity. DUBs's main role is about cleaving specially ubiquitin-linked molecules, which are most of the time a protein. The fate of an ubiquitin-tagged protein depends in part of the length of the ubiquitin chain(s) and on the configuration of ubiquitin-ubiquitin linkages in the chain considered. The tag is going in particular to regulate the degradation of the tagged-molecule by the [https://en.wikipedia.org/wiki/Proteasome proteasome] or [https://en.wikipedia.org/wiki/Lysozyme lysozyme], influence its cellular localisation or modulates the activity with another protein. <ref>PMID:17218518</ref><ref>PMID:12860974</ref> | |
| - | + | ||
| - | == Function == | + | == Generalities == |
| + | |||
| + | ==== Function ==== | ||
| + | |||
| + | Deubiquitinases are key enzymes belonging to the vast group of '''proteases'''.They take part in the removal of ubiquitin molecules on proteins which have been ubiquitinated, allowing the degradation of [https://en.wikipedia.org/wiki/Ubiquitin ubiquitin]-tagged proteins. As a result, these enzymes play an important role being implicated in the regulation of '''protein degradation'''. Indeed, the pathway for a protein to be degraded involves, an enzymatic cascade that will add a poly-ubiquitin fragment to the protein. This mechanism is called [https://fr.wikipedia.org/wiki/Ubiquitination ubiquitination(fr)] [https://en.wikipedia.org/wiki/Ubiquitin#Ubiquitylation (en)]. Following this step, mono or poly-ubiquitin is by deubiquitinases from the protein. | ||
| + | |||
| + | ==== Classes ==== | ||
| + | |||
| + | Deubiquitinases belong to the large protease family. This family gathers into '''five classes''',all of them definded according to the nature of the amino acid composition of their active site carrying out the catalysis. It can be either : serine proteases, [https://en.wikipedia.org/wiki/Cysteine_protease cysteine proteases], acid proteases, metalloproteases, or threonine proteases. DUBs belong to only two of these families: '''metalloproteases''' and '''cysteine proteases'''. | ||
| + | Among the cysteine proteins, four subfamilies can be described according to their catalytic domains: ubiquitin-specific proteases (USPs),the Ubiquitin C-terminal hydrolases (UCHs),Ovarian tumore-related proteases (OTUs) and Machado-Joseph disease proteases (MJD). The deubiquitinases belonging to the family of metalloproteases all have a JAMM catalytic domain (JAB1/MPN/Mov34 metalloenzyme). | ||
| + | Within these two families, DUBs are classified into subfamilies according to the differences in their amino acid sequences surrounding the catalytically active amino acid residues. <ref>PMID:15571815</ref> | ||
| + | |||
| + | ==== Localization ==== | ||
| + | |||
| + | The localization depends on the DUB we consider. Nevertheless, the majority of DUBs are found in the nucleus, plasma membrane and/or in secretory and endocytic pathways. For example, in the ubiquitin-specific proteases family (UPSs), USP21 is mostly associated with microtubules and the [https://en.wikipedia.org/wiki/Centrosome centrosome]. Thus this deubiquitinase is highly dependent on all the physiological mechanisms involving the [https://en.wikipedia.org/wiki/Microtubule microtubules]. <ref>PMID:22298430</ref> | ||
| + | |||
| + | == Structure == | ||
| + | |||
| + | ==== The overall structure ==== | ||
| + | |||
| + | 3TMP is the catalytic domain of human deubiquitinase DUBA in complex with ubiquitin aldehyde. It is a 8-chain-structure with sequences from Human. Indeed, 3TMP is made of two macromolecules : OTU domain-containing protein 5 (also named DUBA or OTUD5) and Polyubiquitin-C, which is the ubiquitin aldehyde. The catalytic domain is also composed of two small molecules the phosphoserine (SEP) and the amino-acetaldehyde (GLZ),which they are L-peptide links. <ref>PMID:22245969</ref> | ||
| + | |||
| + | === Impact of phosphorylation on DUB activity === | ||
| + | |||
| + | Evidence shows that phosphorylation influences enzyme activity. Phosphorylated serine seems to have the most influence on the enzyme activity <scene name='86/868189/Ser177/1'>especially on Ser177</scene>. The phosphorylation of this nucleotide is crucial to the protein to work. | ||
| + | In fact, this part bends to welcome the protein to be deubiquitinated. <ref>PMID:22245969</ref> | ||
| + | |||
| + | ==== Catalytic domain ==== | ||
| + | |||
| + | The DUB family is determined by the catalytic domain. Indeed, DUBs belonging to the family of cysteine proteases have a catalytic site composed of two or three amino acids (dyads or triads). When the catalytic site is active, it may contain cysteine, histidine, aspartate or asparagine residues. As far as the metalloproteases are concerned, their active site is composed of a zinc ion and amino acids such as histidine, aspartate and serine. <ref>https://authors.library.caltech.edu/261/1/AMBpb04.pdf</ref> | ||
| + | The studied structure shows both <scene name='86/868189/Catalytic_site_polyubiquitine/2'>the catalytic site (in orange) and polyubiquitine (in purple).</scene> | ||
| + | |||
| + | Residues present in the catalytic site of DUBs are often in a '''non-functional orientation''' in the absence of substrate. As a result, when the substrate binds to the catalytic site of the enzyme, the site undergoes rearrangements and takes on a functional conformation. <ref>PMID:16537382</ref> The substrate opens and closes to allow the entry of the protein to be deubiquitinased. | ||
| + | The enzyme takes this configuration thanks to <scene name='86/868189/H_bonds_around_ser177/1'>many hydrogen bonds around Ser177.</scene> This is why phosphorylation is so important to the function of the enzyme. The phosphate group forms many links between substrate ubiquitin and a segment of the OTU domain. This is rare among the known structures of deubiquitinases. Phosphorylation-driven conformational change resembles to the one of [https://en.wikipedia.org/wiki/Kinase kinases].<ref>PMID:22245969</ref> | ||
| + | |||
| + | == Biological role == | ||
| + | |||
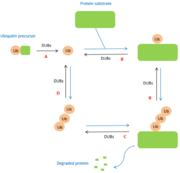
| + | DUBs are involved at multiple levels in the [[ubiquitin]] pathway. The modifications made by DUBs are post-translational modifications. Depending on the level, DUBs have different functions. Two specific cellular functions exist for deubiquitinases. They may act either on the degradation of the stabilization of a substrate in particular <ref>Alan D’Andrea, David Pellman,Deubiquitinating Enzymes: A New Class of Biological Regulators, Taylor and Francis Online, Sept 29 2008,DOI: https://doi.org/10.1080/10409239891204251</ref>. Further functions are more specifically related to the ubiquitin molecule. Here are some of them : | ||
| + | |||
| + | A : '''maturation of ubiquitin'''. When ubiquitin molecules are synthesized, they are not in free form. Thus, DUBs are essential for the generation of free monomers from precursors. The degradation of precursors is carried out by several DUBs belonging to the USPs class. | ||
| + | |||
| + | B : cleavage between protein and mono-ubiquitin and regulation of the poly-ubiquitin chain. DUBs also have a regulatory activity because they allow the elimination of ubiquitin chains mistakenly conjugated to substrates. | ||
| + | |||
| + | C : cleavage between protein and poly-ubiquitin chain. Once the protein has been degraded by the [[proteasome]] or autophagolysosome, DUBs allow the polyubiquitin chain to be separated from the protein. | ||
| + | |||
| + | D : '''recycling of ubiquitin'''. Certain deubiquitinases allow the separation of polyubiquitin chains in order to release ubiquitin monomers. <ref>PMID:15571815</ref> | ||
| + | |||
| + | |||
| + | [[Image:DUBspathways.PNG |thumb|center|Figure 1 : Role of DUBs in the ubiquitin pathways]] | ||
| + | |||
| + | Some other deubiquitinating enzymes also have biological functions. The latter are for instance involved in growth control, transcription silencing, regulation and viral infection or even the processing of ubiquitin-like-modifications.<ref>Alan D’Andrea, David Pellman,Deubiquitinating Enzymes: A New Class of Biological Regulators, Taylor and Francis Online, Sept 29 2008,DOI: https://doi.org/10.1080/10409239891204251</ref> | ||
== Disease == | == Disease == | ||
| - | + | The involvement of deubiquitinases in diseases is still poorly understood. However, we know that they play an important role in various physiological processes, particularly in the case of '''cancers'''. <ref>PMID:19007433</ref> | |
| + | In fact, DUBs have a role in the mechanism involved in [https://en.wikipedia.org/wiki/Histone histone] modification and so have influence on '''tumor development''' and its progression. For instance, in gastric cancer, DUBs are regulated upwards and DUBs are related to tumor size. <ref>PMID:31897112</ref> | ||
| - | == | + | ==== Otubain 1 ==== |
| - | + | The enzyme Otubain 1 is a deubiquitinase belonging to the Otubain family of proteases. The role of OTUB1 is not yet clearly defined, some studies show the correlation between tumor growth and OTUB1 while others show no involvement or suppression of the tumor by this enzyme. This suggests that the effects of OTUB1 depend on the stage and the tumor itself. However, the therapeutic targeting of OTUB1 could be used for patients with various tumors, since the enzyme is found in many tissues and has a high level of cell expression. <ref>PMID:30400005</ref> | |
| + | |||
| + | ==== Treatment ==== | ||
| + | |||
| + | DUBs are attractive targets for drug therapy. As a result, they may be used for it because they are widely involved in key regulatory processes. In fact, DUBs might function to regulate both stability and the activity of target proteins like oncogenes and tumor suppressors.<ref>David Komander, Michael J. Clague, Sylvie Urbé, Breaking the chains: structure and function of the deubiquitinases,Nature reviews molecular cell biology 10, August 2009, DOI:https://doi.org/10.1038/nrm2731</ref> | ||
</StructureSection> | </StructureSection> | ||
| + | |||
== References == | == References == | ||
<references/> | <references/> | ||
Current revision
| This Sandbox is Reserved from 26/11/2020, through 26/11/2021 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1643 through Sandbox Reserved 1664. |
To get started:
More help: Help:Editing |
Deubiquitinases
| |||||||||||
References
- ↑ Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007 Jan 12;315(5809):201-5. doi: 10.1126/science.1127085. PMID:17218518 doi:http://dx.doi.org/10.1126/science.1127085
- ↑ Schnell JD, Hicke L. Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J Biol Chem. 2003 Sep 19;278(38):35857-60. doi: 10.1074/jbc.R300018200. Epub 2003, Jul 14. PMID:12860974 doi:http://dx.doi.org/10.1074/jbc.R300018200
- ↑ Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004 Nov 29;1695(1-3):189-207. doi:, 10.1016/j.bbamcr.2004.10.003. PMID:15571815 doi:http://dx.doi.org/10.1016/j.bbamcr.2004.10.003
- ↑ Urbe S, Liu H, Hayes SD, Heride C, Rigden DJ, Clague MJ. Systematic survey of deubiquitinase localization identifies USP21 as a regulator of centrosome- and microtubule-associated functions. Mol Biol Cell. 2012 Mar;23(6):1095-103. doi: 10.1091/mbc.E11-08-0668. Epub 2012, Feb 1. PMID:22298430 doi:http://dx.doi.org/10.1091/mbc.E11-08-0668
- ↑ Huang OW, Ma X, Yin J, Flinders J, Maurer T, Kayagaki N, Phung Q, Bosanac I, Arnott D, Dixit VM, Hymowitz SG, Starovasnik MA, Cochran AG. Phosphorylation-dependent activity of the deubiquitinase DUBA. Nat Struct Mol Biol. 2012 Jan 15;19(2):171-5. doi: 10.1038/nsmb.2206. PMID:22245969 doi:10.1038/nsmb.2206
- ↑ Huang OW, Ma X, Yin J, Flinders J, Maurer T, Kayagaki N, Phung Q, Bosanac I, Arnott D, Dixit VM, Hymowitz SG, Starovasnik MA, Cochran AG. Phosphorylation-dependent activity of the deubiquitinase DUBA. Nat Struct Mol Biol. 2012 Jan 15;19(2):171-5. doi: 10.1038/nsmb.2206. PMID:22245969 doi:10.1038/nsmb.2206
- ↑ https://authors.library.caltech.edu/261/1/AMBpb04.pdf
- ↑ Das C, Hoang QQ, Kreinbring CA, Luchansky SJ, Meray RK, Ray SS, Lansbury PT, Ringe D, Petsko GA. Structural basis for conformational plasticity of the Parkinson's disease-associated ubiquitin hydrolase UCH-L1. Proc Natl Acad Sci U S A. 2006 Mar 21;103(12):4675-80. Epub 2006 Mar 13. PMID:16537382
- ↑ Huang OW, Ma X, Yin J, Flinders J, Maurer T, Kayagaki N, Phung Q, Bosanac I, Arnott D, Dixit VM, Hymowitz SG, Starovasnik MA, Cochran AG. Phosphorylation-dependent activity of the deubiquitinase DUBA. Nat Struct Mol Biol. 2012 Jan 15;19(2):171-5. doi: 10.1038/nsmb.2206. PMID:22245969 doi:10.1038/nsmb.2206
- ↑ Alan D’Andrea, David Pellman,Deubiquitinating Enzymes: A New Class of Biological Regulators, Taylor and Francis Online, Sept 29 2008,DOI: https://doi.org/10.1080/10409239891204251
- ↑ Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004 Nov 29;1695(1-3):189-207. doi:, 10.1016/j.bbamcr.2004.10.003. PMID:15571815 doi:http://dx.doi.org/10.1016/j.bbamcr.2004.10.003
- ↑ Alan D’Andrea, David Pellman,Deubiquitinating Enzymes: A New Class of Biological Regulators, Taylor and Francis Online, Sept 29 2008,DOI: https://doi.org/10.1080/10409239891204251
- ↑ Singhal S, Taylor MC, Baker RT. Deubiquitylating enzymes and disease. BMC Biochem. 2008 Oct 21;9 Suppl 1:S3. doi: 10.1186/1471-2091-9-S1-S3. PMID:19007433 doi:http://dx.doi.org/10.1186/1471-2091-9-S1-S3
- ↑ Sun J, Shi X, Mamun MAA, Gao Y. The role of deubiquitinating enzymes in gastric cancer. Oncol Lett. 2020 Jan;19(1):30-44. doi: 10.3892/ol.2019.11062. Epub 2019 Nov 7. PMID:31897112 doi:http://dx.doi.org/10.3892/ol.2019.11062
- ↑ Saldana M, VanderVorst K, Berg AL, Lee H, Carraway KL. Otubain 1: a non-canonical deubiquitinase with an emerging role in cancer. Endocr Relat Cancer. 2019 Jan 1;26(1):R1-R14. doi: 10.1530/ERC-18-0264. PMID:30400005 doi:http://dx.doi.org/10.1530/ERC-18-0264
- ↑ David Komander, Michael J. Clague, Sylvie Urbé, Breaking the chains: structure and function of the deubiquitinases,Nature reviews molecular cell biology 10, August 2009, DOI:https://doi.org/10.1038/nrm2731