Sandbox Reserved 1653
From Proteopedia
(Difference between revisions)
| (25 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <references/><StructureSection load='5z10' size='350' side='right' caption='Structure of the mechanosensitive Piezo1 channel 1 from | + | <references/><StructureSection load='5z10' size='350' side='right' caption='Structure of the mechanosensitive Piezo1 channel 1 from [http://www.rcsb.org/structure/5Z10 PBD]' scene=''> |
| - | Piezo1 proteins constitute a family of excitatory [[ion channels]] directly gated by mechanical forces. Piezo1 is functionally conserved and very important because all living organisms are subjected to mechanical forces from their environment for instance [https://en.wikipedia.org/wiki/Proprioception proprioception], [https://en.wikipedia.org/wiki/Osmoregulation osmoregulation], vascular tone, blood flow regulation, muscle [https://en.wikipedia.org/wiki/Homeostasis homeostasis], flow sensing in kidney, bladder and lungs.<ref name="Ion Permeation"> DOI 10.1016/j.neuron.2016.01.046 </ref>,<ref name = " | + | Piezo1 proteins constitute a family of excitatory [[ion channels]] directly gated by mechanical forces. Piezo1 is functionally conserved and very important because all living organisms are subjected to mechanical forces from their environment for instance [https://en.wikipedia.org/wiki/Proprioception proprioception], [https://en.wikipedia.org/wiki/Osmoregulation osmoregulation], vascular tone, blood flow regulation, muscle [https://en.wikipedia.org/wiki/Homeostasis homeostasis], flow sensing in kidney, bladder and lungs.<ref name="Ion Permeation"> DOI 10.1016/j.neuron.2016.01.046 </ref> |
| + | |||
| + | == Structure == | ||
| + | |||
| + | Piezo is a very large protein, approximately 2500 amino acids, that possesses a homotrimeric structure, an intracellular and extracellular domain, and an unusually high number of [https://en.wikipedia.org/wiki/Transmembrane_domain transmembrane domains], up to 40. | ||
| + | |||
| + | ==='''Blade'''=== | ||
| + | |||
| + | Piezo1 has a central domain which is composed of <scene name='86/868186/Cedohihctd_color2/1'>one CTD, one cap (or CED), 3 inner helices (IH) and 3 outer helices (OH)</scene>. | ||
| + | This central domain is surrounded by 3 extended arms called <scene name='86/868186/Blade/2'>blades</scene> extending out from the central pore in a rotatory manner <ref name ="Alexandra"> Zhou, Z. (2019). Structural Analysis of Piezo1 Ion Channel Reveals the Relationship between Amino Acid Sequence Mutations and Human Diseases. 139–155. DOI 10.4236/jbm.2019.712012 </ref>. | ||
| + | "Each of these blades, deflecting at an angle of 100° perpendicular to the membrane, contains 6 tandems transmembranar helical units (THUs) constitute of 4 transmembrane domains".<ref name="mechanogating"/>,<ref name="Alexandra"/> "They are not planar: instead, they lie on a spherically curved surface with the membrane bulging into the cytoplasm".<ref name= "Piezo Senses Tension "> DOI 10.1016/j.cub.2018.02.078</ref> | ||
| + | These flexible blades are inside the membrane and force the membrane to curve. That is why, they are considered as mechanotransduction modules, force sensors and transducers to gate the central pore. "These 3 blades propeller architecture is mechanically interesting because 3 blades are the minimum for omnidirectional sensitivity".<ref name="Piezo Senses Tension "/> | ||
| + | |||
| + | ==='''CED or cap'''=== | ||
| + | |||
| + | The <scene name='86/868186/Ced/1'>CED</scene>(carboxyterminal extracellular domain) also called cap is a large extracellular domain in loop shape that forms a trimer. This CED is located in the central module surrounded by the <scene name='86/868186/Blade/1'>blades</scene> and contains 240 residus.<ref name="Architecture"> DOI 10.1038/nature15247 </ref>,<ref name = "Ion Permeation"/> | ||
| + | This CED mediates efficient ion conduction and cation selectivity because it may allow cations to enter or exit the transmembrane pore. For this, the cap structure may provide a mechanism for enriching cation at the extracellular vestibule by utilizing a large patch of <scene name='86/868186/E_et_d/1'>negatively charged residues (DEEEED)</scene>). | ||
| + | CED constitutes the extracellular <scene name='86/868186/Ion_conducting_pore/1'>ion conducting</scene> pathway to regulate ion permeation as selecticity properties of Piezo1 channels.<ref name = "Ion Permeation"/> | ||
| + | |||
| + | ==='''Ion conducting pore'''=== | ||
| + | The <scene name='86/868186/Ion_conducting_pore/1'>central pore axis</scene> of Piezo1 is lined with the <scene name='86/868186/Ced/1'>extracellular cap domain</scene>, inner helix and cytosolic <scene name='86/868186/Ctd/1'>CTD</scene>. The extracellular cations can approach the pore entry “vertically through the internal cavity along the threefold axis of the cap domain”, they can also approach laterally through spaces (gaps) between the flexible linkers which connect the cap with inner and outer helices.<ref name= "ion channel"> DOI 10.1038/nature25453 </ref> The <scene name='86/868186/Ion_conducting_pore/1'>ion conduction pathway</scene> is situated below the <scene name='86/868186/Ced/1'>cap</scene>, and is lined by the three inner transmembrane helices. The possible access for lipids or other hydrophobic molecules through the pore could be “two lateral openings between the inner helices separated by a ‘seal’ formed by <scene name='86/868186/K2479_f2480/1'>K2479 and F2480</scene>”. These openings are approximately 11 Å wide and 16 Å tall.<ref name= "ion channel"/> | ||
| + | |||
| + | ==='''CTD and Beam'''=== | ||
| + | |||
| + | CTD and <scene name='86/868186/Beam_1_couleurs/1'>beams</scene> are intracellular. The beam interacts with the CTD, and both are required for mechanical activation of the channel.<ref name="Architecture"/> | ||
| + | |||
| + | The <scene name='86/868186/Ctd_trimeric/1'>CTD</scene> is a trimeric structure and is a part of the pore module of Piezo1 channel. The CTD interacts with the long <scene name='86/868186/Anchor/1'>anchorα</scene>, and forms a hydrophobic interface. This forms a tripartite interaction with the <scene name='86/868186/E_dans_ctd/1'>glutamate-rich regions of the CTD</scene>.<ref name="mechanogating"> DOI 10.1038/nature25743 </ref> | ||
| + | It forms an intracellular vestibule along the z-axis, and it is essential for ion permeation properties.<ref name="Architecture"> DOI 10.1038/nature15247 </ref> The CTD triangular plane has a beam-facing side of the triangular, and it is separated into two surfaces with negative and positive electrostatic potentials.<ref name="mechanogating"/> | ||
| + | |||
| + | The beam is a 90 Å-long intracellular structure in the central region of the ion channel. It is a <scene name='86/868186/Blade_ans_beam/1'>part of the three-bladed</scene>, propeller-shaped architecture characteristic of Piezo1. It is a piece of the “beam-CTD-anchor-OH-IH” relaying interface that forms the central pore module. It is because the beam connects the THU, to the CTD and the outer helix (OH) that it enables the transmission of the mechanical force, and thus the opening of Piezo1’s pore.<ref name="mechanogating"/> It delivers the mechanical signals from the blades, or the plasma membrane, to the central pore module region.<ref name="Alexandra"/> | ||
| + | Indeed, the beams are connected to the transmembrane helical units (THUs), which forms a triangular plane above its proximal end. This makes the largest intracellular loop of Piezo1, and it starts at the distal end of the beam, interacts with the CTD, and then folds back to the distal end of the beam before connecting to a transmembrane region. Moreover, the beam crosses through the beam-facing side of the triangular CTD, forming interactions with both CTDα 1 and CTDα 2. | ||
| + | This position and connections of the beam render it an ideal structure for mechanical transmission from the distal THUs to the central ion-conducting pore. The lever-like mechanotransduction apparatus constituted by the beam is possible because of its uneven movement. It displays large motion at the distal beam while subtle movement at the proximal end. It enables Piezo1 channels to effectively convert a large conformational change of the distal blades to a relatively slight opening of the central pore, allowing cation-selective permeation.<ref name="mechanogating"/> | ||
| + | |||
| + | |||
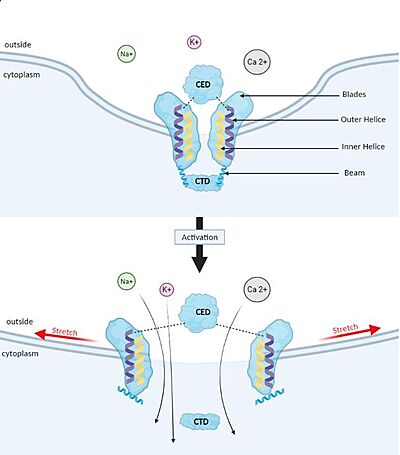
| + | [[Image:Piezo_1_opening_mechanism.JPG#filehistory | thumb |left|400px| upright=10| '''Membrane deformation and Gating mechanism of Piezo1 before and after activation by an external mechanical force.''']] | ||
| + | ==='''Gating mechanism'''=== | ||
| + | Piezo1 possesses delicate force sensing and mechanotransduction mechanisms. Here, we explain how Piezo1 channels sense and transduce mechanical forces to gate the central ion-conducting pore. | ||
| + | Piezo1 can sense membrane tension through changes in the local curvature of the membrane and the channel opens in response to this change thanks to this structure.<ref name ="Piezo Senses Tension"/> | ||
| + | Indeed, mPiezo trimer is a non-planar conformation inside lipid bilayer, it produces a local dome-shaped deformation of the membrane. In cells, this membrane curvature project towards the cytoplasm and some electrostatic interactions stabilize the trimeric assembly in its curved conformation.<ref name = "nv article"> DOI 10.7554/eLife.33660</ref> | ||
| + | The structure of Piezo1 offers a plausible explanation for the origin of its tension [https://en.wikipedia.org/wiki/Gating_(electrophysiology) gating]. Indeed, if the semi-spherical dome becomes flatter when Piezo1 opens, then the channel membrane system will expand thanks to the flexibility of the blades. | ||
| + | However, because flattening does not constrain the pore to open wide, expansion and pore diameter are decoupled, such that Piezo1 can exhibit its small conductance and cation selectivity, properties that are essential to its function.<ref name ="Piezo Senses Tension"/>,<ref name="Lin" />. Despite the recent functional evidence, our understanding is limited due to the lack of knowledge on the N-term domain and the cytoplasmic loops of Piezo1. However, recently a modelization of the full length of Piezo 1 confirmed the above-proposed mechanism, showed the importance of the N-terminal domain in shaping the topology of the membrane surrounding Piezo1, and suggested the implication of the cytoplasmic loop as a contact site with the cytoskeleton or as a site for post-translational modification <ref name = "Yoda1"> DOI 10.1016/j.bpj.2021.02.003 </ref>. | ||
| + | |||
== Function == | == Function == | ||
| Line 15: | Line 56: | ||
Cells are able to perceive the filling of the stomach or the bladder, blood flow and lungs inflation. | Cells are able to perceive the filling of the stomach or the bladder, blood flow and lungs inflation. | ||
| - | Piezo1 is a sensor of mechanical forces in [https://en.wikipedia.org/wiki/Endothelium endothelial], urothelial and renal epithelial cells. For instance, Piezo1 is involved in shear stress sensing in blood vessel endothelial cells and is implicated in the development and physiological functions of the circulatory system, including the proper formation of blood, vessels, regulation of vascular tone and remodelling of small resistance arteries upon [https://en.wikipedia.org/wiki/Hypertension hypertension]. It is also involved in the homeostasis of the volume of red blood cells. | + | Piezo1 is a sensor of mechanical forces in [https://en.wikipedia.org/wiki/Endothelium endothelial], urothelial and renal epithelial cells. For instance, Piezo1 is involved in shear stress sensing in blood vessel endothelial cells and is implicated in the development and physiological functions of the circulatory system, including the proper formation of blood, vessels, regulation of vascular tone and remodelling of small resistance arteries upon [https://en.wikipedia.org/wiki/Hypertension hypertension]. It is also involved in the homeostasis of the volume of red blood cells. |
Piezo1 mediates cationic non-selective currents. Indeed, monovalent (Na+, K+) and divalent (Ca2+, Mg2+) can flow through Piezo1. | Piezo1 mediates cationic non-selective currents. Indeed, monovalent (Na+, K+) and divalent (Ca2+, Mg2+) can flow through Piezo1. | ||
However, Piezo1 is implicated in excitatory channels because cations can enter into the cells which leads to the membrane [https://en.wikipedia.org/wiki/Depolarization depolarisation] or [https://en.wikipedia.org/wiki/Calcium_signaling calcium-dependent signalling pathway] (if Ca2+ enters).<ref name="Adenosine"> DOI 10.3389/fphar.2019.01304 </ref> | However, Piezo1 is implicated in excitatory channels because cations can enter into the cells which leads to the membrane [https://en.wikipedia.org/wiki/Depolarization depolarisation] or [https://en.wikipedia.org/wiki/Calcium_signaling calcium-dependent signalling pathway] (if Ca2+ enters).<ref name="Adenosine"> DOI 10.3389/fphar.2019.01304 </ref> | ||
| Line 29: | Line 70: | ||
| - | == | + | == Diseases == |
| - | + | Since Piezo1 is implicated in the functioning of many cells and organs, modifications on its structure lead to diseases. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | For instance, Hereditary Xerocytosis (HX) is a rare disease, also called [https://en.wikipedia.org/wiki/Hereditary_stomatocytosis Dehydrated hereditary stomatocytosis] (DHS). This genetic disease leads to impaired red blood cell (RBC) membrane properties that affect intracellular cation concentrations.<ref name="Dehydrated"> DOI 10.1038/ncomms2899</ref> The RBCs are abnormally shaped and they result in [https://en.wikipedia.org/wiki/Hemolytic_anemia haemolytic anaemia]. Those modifications are due to mutations on the FAM38A gene on chromosome 16 which encodes for Piezo 1. Piezo1 is expressed in the plasma membranes of RBCs, and its role is to control RBCs’ osmolarity. It also plays a prevalent role in the [https://en.wikipedia.org/wiki/Erythropoiesis erythroid differentiation]. Mutations in Piezo1 distort mechanosensitive channel regulation, leading to increased cation transport in erythroid cells. Those mutations affect different parts of the channel. For instance, six gain-of-function mutations, gathered in the central core region of the Piezo channel structure, are directly linked to the decrease of inactivation rate of the channel. Mutations in the N-terminal part of the protein also have a role in channel gating. Therefore, not every channel is affected in the same way and by the same mutation. Indeed it depends on the environment, the permeability of RBC and the combination of mutations <ref name = "HX"> DOI 10.3389/fphys.2021.736585 </ref>. | ||
| + | Those mutations could provoke increases in permeability of cations in RBC by different mechanisms. It could induce mechanically activated currents that inactivate more slowly than wild-type currents. They could also affect the inactivation process by either destabilising the inactivated state or stabilising the channel in the open state. As a result, the open to inactivated state equilibrium shifts towards open. Na+ and Ca2+ ion influx consequently increase, and the intracellular K+ concentration decreases in a steady state. The evolution of Piezo1’s function steams from a change in its 3D structure. | ||
| + | Lymphatic dysplasia <ref name = "Lymphatic dysplasia"> DOI 10.1016/bs.ctm.2017.01.001 </ref> is also a disease linked to loss of function mutations on Piezo1. The [https://en.wikipedia.org/wiki/Lymphatic_system lymphatic system] is independent from the vascular one, and its role is to transport antigens responsible for the immune response. If the interstitial fluid is not drained correctly back to the blood, it leads to local inflammation. The mutations on Piezo1 inactivate the channel gate and in this case the concentration of calcium is not increased. The protein isn’t sensitive to the pressure anymore. | ||
| - | ==='''Blade'''=== | ||
| - | + | == Potential therapeutic target == | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | = | + | Piezo1 is a protein discovered recently. Therefore, its potential in medicine is in constant evolution. |
| + | Piezo1 can be used as diagnostic [https://en.wikipedia.org/wiki/Biomarker biomarkers] because it detects different mechanical forces and triggers a specific response adapted to the changes in the environment. For instance, it is sensitive to shear stress which has a major role in [https://en.wikipedia.org/wiki/Cardiovascular_physiology cardiovascular physiology]<ref name = "Atherosclerosis"> DOI 10.2147/JIR.S319789 </ref>. | ||
| + | This mechanosensitive receptor is a candidate for therapeutic innovation more specifically in the case of cardiovascular and neurodegenerative diseases. | ||
| + | These new therapies could be based on Piezo1 pharmacological modulators. Recently, several modulators have been found. | ||
| - | + | - [https://en.wikipedia.org/wiki/Receptor_antagonist Antagonists] like peptide GsMTx4 <ref name = "GsMTx4"> DOI 10.1002/glia.23722 </ref> prevent the induction of [https://en.wikipedia.org/wiki/Demyelinating_disease demyelination], playing therefore a neuroprotective role. This peptide is an inhibitor of Piezo1. | |
| - | + | ||
| - | + | ||
| - | + | - [https://en.wikipedia.org/wiki/Agonist Agonists] like [https://en.wikipedia.org/wiki/Yoda1 Yoda1] <ref name = "Yoda1"> DOI 10.1016/j.bbrc.2019.04.139 </ref> (a synthetic small molecule) enhance channels opening leading to demyelination and damaging the central nervous system. This molecule can activate the Piezo1 channel without mechanical stimulation. However, it can be useful to suppress the migration of transformed cells like [https://en.wikipedia.org/wiki/Fibroblast fibroblasts]. [https://en.wikipedia.org/wiki/Jedi2 Jedi2] is another chemical activator of Piezo1, but it doesn’t act on the same site as Yoda1. | |
| - | + | ||
| - | = | + | - Tubeimoside 1 (TBMS1) is an inhibitor of Yoda1 allowing a decrease of the activity of Piezo 1 channels in endothelial cells. The mechanism is still unclear but according to studies <ref name = "Tubeimoside"> DOI 10.3389/fphar.2020.00768 </ref>, TBMS1 might be a [https://en.wikipedia.org/wiki/Competitive_inhibition competitive inhibitor], meaning that it fixes itself on the same binding site as Yoda1 and is specific to Piezo1 channels. |
| - | + | Discovering those modulators allows a better understanding of the mechanisms of Piezo1 channels and highlights its usefulness in the pharmacological field. | |
| + | |||
| - | + | == Relevance == | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | A majority of Piezo1’s structure has been resolved by [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy cryo-em] but still information is lacking on the N-term domain and some subregions. There is no full structure available yet which can be troublesome to understand the mechanism of the protein. Moreover, Piezo1 is present in a large number of tissues, but the differences in its roles are unclear. Further research is required to allow a better understanding of diseases linked to Piezo1 (like DHS), and thus a better treatment of those diseases. | ||
| - | + | [http://www.rcsb.org/structure/5Z10 PBD] | |
| - | [ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</StructureSection> | </StructureSection> | ||
Current revision
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Zhao Q, Wu K, Geng J, Chi S, Wang Y, Zhi P, Zhang M, Xiao B. Ion Permeation and Mechanotransduction Mechanisms of Mechanosensitive Piezo Channels. Neuron. 2016 Mar 16;89(6):1248-1263. doi: 10.1016/j.neuron.2016.01.046. Epub 2016, Feb 25. PMID:26924440 doi:http://dx.doi.org/10.1016/j.neuron.2016.01.046
- ↑ 2.0 2.1 2.2 Zhou, Z. (2019). Structural Analysis of Piezo1 Ion Channel Reveals the Relationship between Amino Acid Sequence Mutations and Human Diseases. 139–155. DOI 10.4236/jbm.2019.712012
- ↑ 3.0 3.1 3.2 3.3 3.4 Zhao Q, Zhou H, Chi S, Wang Y, Wang J, Geng J, Wu K, Liu W, Zhang T, Dong MQ, Wang J, Li X, Xiao B. Structure and mechanogating mechanism of the Piezo1 channel. Nature. 2018 Feb 22;554(7693):487-492. doi: 10.1038/nature25743. Epub 2018 Jan, 22. PMID:29469092 doi:http://dx.doi.org/10.1038/nature25743
- ↑ 4.0 4.1 4.2 4.3 Liang X, Howard J. Structural Biology: Piezo Senses Tension through Curvature. Curr Biol. 2018 Apr 23;28(8):R357-R359. doi: 10.1016/j.cub.2018.02.078. PMID:29689211 doi:http://dx.doi.org/10.1016/j.cub.2018.02.078
- ↑ 5.0 5.1 5.2 Ge J, Li W, Zhao Q, Li N, Chen M, Zhi P, Li R, Gao N, Xiao B, Yang M. Architecture of the mammalian mechanosensitive Piezo1 channel. Nature. 2015 Nov 5;527(7576):64-9. doi: 10.1038/nature15247. Epub 2015 Sep 21. PMID:26390154 doi:http://dx.doi.org/10.1038/nature15247
- ↑ 6.0 6.1 Saotome K, Murthy SE, Kefauver JM, Whitwam T, Patapoutian A, Ward AB. Structure of the mechanically activated ion channel Piezo1. Nature. 2017 Dec 20. pii: nature25453. doi: 10.1038/nature25453. PMID:29261642 doi:http://dx.doi.org/10.1038/nature25453
- ↑ Guo YR, MacKinnon R. Structure-based membrane dome mechanism for Piezo mechanosensitivity. Elife. 2017 Dec 12;6. pii: 33660. doi: 10.7554/eLife.33660. PMID:29231809 doi:http://dx.doi.org/10.7554/eLife.33660
- ↑ 8.0 8.1 Lin YC, Guo YR, Miyagi A, Levring J, MacKinnon R, Scheuring S. Force-induced conformational changes in PIEZO1. Nature. 2019 Sep;573(7773):230-234. doi: 10.1038/s41586-019-1499-2. Epub 2019 Aug, 21. PMID:31435018 doi:http://dx.doi.org/10.1038/s41586-019-1499-2
- ↑ 9.0 9.1 Chong J, De Vecchis D, Hyman AJ, Povstyan OV, Ludlow MJ, Shi J, Beech DJ, Kalli AC. Modeling of full-length Piezo1 suggests importance of the proximal N-terminus for dome structure. Biophys J. 2021 Apr 20;120(8):1343-1356. doi: 10.1016/j.bpj.2021.02.003. Epub, 2021 Feb 12. PMID:33582137 doi:http://dx.doi.org/10.1016/j.bpj.2021.02.003
- ↑ 10.0 10.1 10.2 10.3 10.4 Wei L, Mousawi F, Li D, Roger S, Li J, Yang X, Jiang LH. Adenosine Triphosphate Release and P2 Receptor Signaling in Piezo1 Channel-Dependent Mechanoregulation. Front Pharmacol. 2019 Nov 6;10:1304. doi: 10.3389/fphar.2019.01304. eCollection, 2019. PMID:31780935 doi:http://dx.doi.org/10.3389/fphar.2019.01304
- ↑ Li J, Hou B, Tumova S, Muraki K, Bruns A, Ludlow MJ, Sedo A, Hyman AJ, McKeown L, Young RS, Yuldasheva NY, Majeed Y, Wilson LA, Rode B, Bailey MA, Kim HR, Fu Z, Carter DA, Bilton J, Imrie H, Ajuh P, Dear TN, Cubbon RM, Kearney MT, Prasad KR, Evans PC, Ainscough JF, Beech DJ. Piezo1 integration of vascular architecture with physiological force. Nature. 2014 Nov 13;515(7526):279-82. doi: 10.1038/nature13701. Epub 2014 Aug 10. PMID:25119035 doi:http://dx.doi.org/10.1038/nature13701
- ↑ Albuisson J, Murthy SE, Bandell M, Coste B, Louis-Dit-Picard H, Mathur J, Feneant-Thibault M, Tertian G, de Jaureguiberry JP, Syfuss PY, Cahalan S, Garcon L, Toutain F, Simon Rohrlich P, Delaunay J, Picard V, Jeunemaitre X, Patapoutian A. Dehydrated hereditary stomatocytosis linked to gain-of-function mutations in mechanically activated PIEZO1 ion channels. Nat Commun. 2013;4:1884. doi: 10.1038/ncomms2899. PMID:23695678 doi:http://dx.doi.org/10.1038/ncomms2899
- ↑ Yamaguchi Y, Allegrini B, Rapetti-Mauss R, Picard V, Garcon L, Kohl P, Soriani O, Peyronnet R, Guizouarn H. Hereditary Xerocytosis: Differential Behavior of PIEZO1 Mutations in the N-Terminal Extracellular Domain Between Red Blood Cells and HEK Cells. Front Physiol. 2021 Oct 18;12:736585. doi: 10.3389/fphys.2021.736585. eCollection, 2021. PMID:34737711 doi:http://dx.doi.org/10.3389/fphys.2021.736585
- ↑ Alper SL. Genetic Diseases of PIEZO1 and PIEZO2 Dysfunction. Curr Top Membr. 2017;79:97-134. doi: 10.1016/bs.ctm.2017.01.001. Epub 2017 Feb, 28. PMID:28728825 doi:http://dx.doi.org/10.1016/bs.ctm.2017.01.001
- ↑ Shinge SAU, Zhang D, Achu Muluh T, Nie Y, Yu F. Mechanosensitive Piezo1 Channel Evoked-Mechanical Signals in Atherosclerosis. J Inflamm Res. 2021 Jul 27;14:3621-3636. doi: 10.2147/JIR.S319789. eCollection, 2021. PMID:34349540 doi:http://dx.doi.org/10.2147/JIR.S319789
- ↑ Velasco-Estevez M, Gadalla KKE, Linan-Barba N, Cobb S, Dev KK, Sheridan GK. Inhibition of Piezo1 attenuates demyelination in the central nervous system. Glia. 2020 Feb;68(2):356-375. doi: 10.1002/glia.23722. Epub 2019 Oct 9. PMID:31596529 doi:http://dx.doi.org/10.1002/glia.23722
- ↑ Liu S, Pan X, Cheng W, Deng B, He Y, Zhang L, Ning Y, Li J. Tubeimoside I Antagonizes Yoda1-Evoked Piezo1 Channel Activation. Front Pharmacol. 2020 May 25;11:768. doi: 10.3389/fphar.2020.00768. eCollection, 2020. PMID:32523536 doi:http://dx.doi.org/10.3389/fphar.2020.00768