We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Abbey Wells/Sandbox 1
From Proteopedia
< User:Abbey Wells(Difference between revisions)
| (35 intermediate revisions not shown.) | |||
| Line 2: | Line 2: | ||
<StructureSection load='6WF2' size='350' frame = 'true' side='right' caption='Structure of SCD1' scene='87/877606/Introduction-di-iron-image/1'> | <StructureSection load='6WF2' size='350' frame = 'true' side='right' caption='Structure of SCD1' scene='87/877606/Introduction-di-iron-image/1'> | ||
| - | ==Introduction== | + | == Introduction == |
| - | + | Stearoyl-CoA Desaturase is an enzyme essential for the biosynthesis of monosaturated fatty acids from saturated fatty acids <ref name="Paton">doi: 10.1152/ajpendo.90897.2008</ref>. SCD catalyzes the rate-limiting step in the conversion of [http://en.wikipedia.org/wiki/Stearoyl-CoA Stearoyl-CoA] to [http://en.wikipedia.org/wiki/Oleic_acid oleic acid], an essential substrate in the biosynthesis of phospholipids, triacyclglycerols, and cholesterol <ref name="Bai">PMID:26098370</ref>. SCD is highly conserved in eukaryotes and has different gene isoforms. Mice have four isoforms: SCD1, SCD2, SCD3, and SCD4. Humans have two different isoforms: SCD1 and SCD5 <ref name="Bai">PMID:26098370</ref>. The SCD isoform discussed in this page is [http://en.wikipedia.org/wiki/Stearoyl-CoA_desaturase-1 Stearoyl-CoA Desaturase 1 (SCD1)] found in mice. SCD is believed to have once been an anaerobic pathway found in cartilaginous fish about 450 million years ago <ref name="Filipe">doi: 10.1186/1471-2148-11-132</ref>. The enzyme’s mechanism is now aerobic and favorable. The structure of SCD1 was found using X-ray crystallography <ref name="Bai">PMID:26098370</ref>. | |
| - | + | ||
| - | + | ||
== Structure == | == Structure == | ||
=== Overall Structure === | === Overall Structure === | ||
| - | SCD1 is embedded within | + | SCD1 is an integral membrane protein embedded within the [http://micro.magnet.fsu.edu/cells/endoplasmicreticulum/endoplasmicreticulum.html endoplasmic reticulum] and consists of 4 transmembrane alpha helices, arranged in a cone-like shape. The cytosolic domain of the enzyme consists of 11 alpha helices and contains the carboxy and amino termini <ref name="Bai">PMID:26098370</ref>. Its substrate, Stearoyl-CoA, binds to the cytosolic region which contains a "kink" that properly orients Stearoyl-CoA to undergo a [http://en.wikipedia.org/wiki/Dehydrogenation#:~:text=Dehydrogenation%20is%20the%20a%20chemical,reaction%20and%20a%20serious%20problem.&text=Enzymes%20that%20catalyze%20dehydrogenation%20are%20called%20dehydrogenases. dehydrogenation] reaction between the <scene name='87/877602/C9_and_c103'>carbon 9 and carbon 10</scene> of Stearoyl-CoA <ref name="Bai">PMID:26098370</ref>. |
| - | + | === Binding of Substrate === | |
| + | [http://en.wikipedia.org/wiki/Stearoyl-CoA Stearoyl-CoA] is the substrate that binds to the enzyme, SCD1. The binding of the substrate is stabilized by specific residues on the exterior and interior of the protein. Stearoyl-CoA is a long-chain fatty [http://en.wikipedia.org/wiki/Acyl-CoA#:~:text=Acyl%2DCoA%20is%20a%20group,forming%20several%20equivalents%20of%20ATP. acyl-CoA]. The head group of the substrate is composed of an adenine, ribose, phosphate groups, and polar atoms such as of nitrogen, oxygen, and sulfur. The head of stearoyl-CoA is attached to the exterior of the protein by polar residues. The adenine, ribose, and phosphate are attached by the residues <scene name='87/877602/Hydrophillic_top_t/4'>R151, D152, K185</scene>. The remaining exterior of the substrate is attached by the residues <scene name='87/877602/Hydrophillic_bottom_labeled_t/5'>N144, N71, R184</scene> <ref name="Bai">PMID:26098370</ref>. All the conserved residues are attached to the Stearoyl-CoA via hydrogen bonds. The fatty acid tail of Stearoyl-CoA is a 17-carbon chain which reaches into the interior of the protein. The fatty acid chain dives into the interior hydrophobic tunnel which is long, narrow, and approximately 24 Angstroms long <ref name="Bai">PMID:26098370</ref>. The geometry of the tunnel and formation of bound acyl chain are the structural basis for the stereospecificity of the desaturation reaction <ref name="Bai">PMID:26098370</ref>. | ||
| + | === Kink of Substrate === | ||
| + | The chain is kinked at <scene name='87/877602/C9_and_c10/3'>carbon 9 and carbon 10</scene> where the double bond is generated. The kink is induced through the interactions of four conserved residues. Three out of four of these residues are not bound to the chain, but are hydrogen bonded to each other: <scene name='87/877602/Kink_build/4'>T257, Q143, W149</scene>. T257 is hydrogen bonded to Q143, and Q143 is hydrogen bonded to W149 <ref name="Bai">PMID:26098370</ref>. These residues are directly below the kink and will be hydrolyzed when the enzymatic product is ready to be released. Specifically, if the hydrogen bond between T257 and Q143 is broken, a large opening would allow for the product to be released into the bilayer <ref name="Bai">PMID:26098370</ref>. The residue that is directly hydrogen bonded to the chain is <scene name='87/877602/W258/4'>W258</scene>. This residue is highly conserved and stabilizes the chain so it will be in the correct orientation in the active site. The enzyme will be effective on acyl chains that are between 17 to 19 carbons long. The residue that has a role in determining substrate length is <scene name='87/877602/Cap/5'>Y104</scene>. Y104 is a capping residue that has approximately 4 [http://en.wikipedia.org/wiki/Angstrom Angstroms] between its' hydroxyl oxygen and the end of the chain <ref name="Bai">PMID:26098370</ref>. | ||
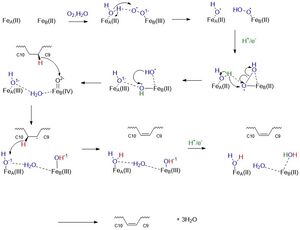
| - | Mechanistically, this reaction involves a molecular oxygen, water molecule, and the transport of electrons down an [http://en.wikipedia.org/wiki/Electron_transport_chain electron transport chain] consisting of cytochrome b5 reductase, cytochrome b5, and NADH to the irons ions within SCD1 which then through a series of redox reactions introduces a double bond between the 9th and 10th carbons of Stearoyl-CoA forming oleic acid. | ||
| - | |||
| - | === Binding of Substrate === | ||
| - | [http://en.wikipedia.org/wiki/Stearoyl-CoA Stearoyl-CoA] is the substrate that binds to the enzyme, SCD1. The binding of the substrate is stabilized by specific residues on the exterior and interior of the protein. Stearoyl-CoA is a long-chain fatty acyl-CoA. The head group of the substrate is composed of an adenine, ribose, phosphate groups, and [http://en.wikipedia.org/wiki/Chemical_polarity#:~:text=In%20chemistry%2C%20polarity%20is%20a,electronegativity%20between%20the%20bonded%20atoms polar atoms] such as of nitrogen, oxygen, and sulfur. The fatty acid tail is a 17-carbon chain which reaches into the interior of the protein. The head of stearoyl-CoA is attached to the exterior of the protein by polar residues. The adenine, ribose, and phosphate are attached by the residues <scene name='87/877602/Hydrophillic_top/2'>R151, D152, K185</scene>. The rest of the exterior of the substrate is attached by the residues <scene name='87/877602/Hydrophillic_bottom_labeled/2'>R184, N144, N71</scene> . The fatty acid chain dives into the interior of the enzyme. The acyl chain is enclosed in a tunnel (24 ang) buried in the cytosolic domain. The geometry of the tunnel and formation of bound acyl chain are the structural basis for the stereospecificity of the desaturation reaction <ref name="Bai">PMID:26098370</ref>. | ||
| - | === Kink of Chain === | ||
| - | The chain is kinked at <scene name='87/877602/C9_and_c10/1'>Carbon 9 and Carbon 10</scene> where the double bond is generated. The Kink is induced through the interactions of four conserved residues. Three out of four of these residues are not bound to the chain, but are hydrogen bonded to each other: <scene name='87/877602/Kink_build/1'>T257, Q143, W149</scene>. T257 is hydrogen bonded to Q143, and Q143 is hydrogen bonded to W149. These residues are directly below the kink and will be hydrolyzed when the substrate is ready to be released. The residue that is directly hydrogen bonded to the chain is <scene name='87/877602/W258/1'>W258</scene>. This residue is highly conserved and stabilizes the chain so it will be in the correct orientation in the active site. | ||
| - | <scene name='87/877602/C9_and_c10/1'>Y104</scene> | ||
=== Active Site === | === Active Site === | ||
| - | Two structures of SCD known. One structure shows the substrate, water molecule, and zinc (4YMK). The second structure shows the product and iron (6WF2). For the pictures below, the structure is shown with two zinc ions in order to demonstrate how is <scene name='87/877627/Zn_and_water/1'>water</scene> coordination with the ions. Water is used in the mechanism, which is why we chose to use the structure with zinc to explain the active site. | ||
| - | The two <scene name='87/877627/Just_zn/1'>zinc</scene>ions are 6.4 Angstroms apart. The ions sit above the kink in the active site. They are stabilized by the <scene name='87/877627/His_box2/1'>his box</scene>. The ion closest to C9 is 5.2 angstroms from it. This ion interacts with 4 histidines, H156, H265, H294, H298, and one water molecule. The ion closest to C10 is 4.7 angstroms away from it. This ion interacts with 5 histidines, H116, H121, H153, H157, and H297. These 9 total Histidine residues form a <scene name='87/877627/His_box2/1'>his box</scene>.The his box is used to stabilize the ions into the active site, forming a prosthetic group. The his box is highly conserved among the other isoforms of SCD. Other residues around the his box are used to hydrogen bond to the histidines to stabilize them. These residues include: <scene name='87/877627/E161/1'>E161,</scene> | ||
| - | <scene name='87/877627/N261/1'>N261,</scene> <scene name='87/877627/D165/1'>D165,</scene> <scene name='87/877627/E291/1'>and E291</scene> | ||
| Line 28: | Line 19: | ||
[[Image:SCDMech.jpg|300px|thumb|left|]] | [[Image:SCDMech.jpg|300px|thumb|left|]] | ||
== Disease == | == Disease == | ||
| - | One common mutation that leads to the loss of function SCD1 is a mutation at any of the histidine residues within the <scene name='87/877627/His_box2/1'>his box.</scene> A mutation at any of these 9 histidine residues interrupts the active site. This knockout of SCD1 has been known to combat obesity and lead to liver disease. | ||
| - | Another common mutation with SCD is the insertion of a proline at position <scene name='87/877627/R279/1'>279.</scene> This is due to an insertion of a 'CCC' codon at position 835 in exon 5 of the SCD1 gene. This mutation results in a loss of function of SCD1. This study was done using a mouse model. In mice with this mutation, hair loss, similar to alopecia, occurs. The mice were also found to be lean throughout their lifespan due to reduced triglyceride synthesis due to loss of SCD1 function. | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<ref name="Bai">PMID:26098370</ref> | <ref name="Bai">PMID:26098370</ref> | ||
<ref name="Shen">PMID:32470559</ref> | <ref name="Shen">PMID:32470559</ref> | ||
| + | <ref name="Filipe">doi: 10.1186/1471-2148-11-132</ref> | ||
| + | <ref name="Paton">doi: 10.1152/ajpendo.90897.2008</ref> | ||
<references/> | <references/> | ||
== Student Contributors == | == Student Contributors == | ||
Current revision
Stearoyl-CoA Desaturase 1 from Mus musculus
| |||||||||||
References
- ↑ 1.0 1.1 Paton CM, Ntambi JM. Biochemical and physiological function of stearoyl-CoA desaturase. Am J Physiol Endocrinol Metab. 2009 Jul;297(1):E28-37. doi:, 10.1152/ajpendo.90897.2008. Epub 2008 Dec 9. PMID:19066317 doi:http://dx.doi.org/10.1152/ajpendo.90897.2008
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 Bai Y, McCoy JG, Levin EJ, Sobrado P, Rajashankar KR, Fox BG, Zhou M. X-ray structure of a mammalian stearoyl-CoA desaturase. Nature. 2015 Jun 22. doi: 10.1038/nature14549. PMID:26098370 doi:http://dx.doi.org/10.1038/nature14549
- ↑ 3.0 3.1 Castro LF, Wilson JM, Goncalves O, Galante-Oliveira S, Rocha E, Cunha I. The evolutionary history of the stearoyl-CoA desaturase gene family in vertebrates. BMC Evol Biol. 2011 May 19;11:132. doi: 10.1186/1471-2148-11-132. PMID:21595943 doi:http://dx.doi.org/10.1186/1471-2148-11-132
- ↑ Shen J, Wu G, Tsai AL, Zhou M. Structure and Mechanism of a Unique Diiron Center in Mammalian Stearoyl-CoA Desaturase. J Mol Biol. 2020 May 27. pii: S0022-2836(20)30367-3. doi:, 10.1016/j.jmb.2020.05.017. PMID:32470559 doi:http://dx.doi.org/10.1016/j.jmb.2020.05.017
Student Contributors
- Abbey Wells
- Josey McKinley
- Anthony Durand