Sandbox Reserved 1659
From Proteopedia
(Difference between revisions)
| (One intermediate revision not shown.) | |||
| Line 19: | Line 19: | ||
== Structure== | == Structure== | ||
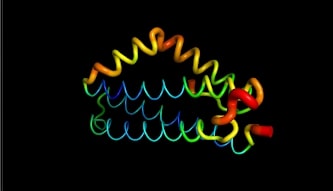
LntA is a small basic protein of 9.7 kDa. This protein is highly conserved in L. monocytogenes and is absent in almost all non-pathogenic Listeria strains . This characteristic suggests that lntA plays a key role in Listeria’s virulence. The acidic part of LntA is composed of aspartic acid (<scene name='86/868192/Acidic/1'>17,8%</scene>) and the basic part is composed of lysine and arginine (<scene name='86/868192/Basic/1'>18,6%</scene>). LntA folds in a compact helical structure and is composed of 5 alpha-helix, three of them are long antiparallel helix and can be seen as the core of the protein. The two remaining helix stick out the core. The 3 first helix are named <scene name='86/868192/Helix_h1/1'>H1</scene>, <scene name='86/868192/Helix_h2/1'>H2</scene> and <scene name='86/868192/Helix_h3/1'>H3</scene>. The 2 others are <scene name='86/868192/Helix_h4ter/1'>H4</scene> and <scene name='86/868192/Helix_h5/4'>H5</scene>. These residues located in <scene name='86/868192/These_two_helix/1'>these two helixes</scene> display high RMSD values meaning that this region is likely to oscillate. In fact, by studying this protein on pymol we can see it by the thickness and redness of these helixes which means that they have a high RMSD value : | LntA is a small basic protein of 9.7 kDa. This protein is highly conserved in L. monocytogenes and is absent in almost all non-pathogenic Listeria strains . This characteristic suggests that lntA plays a key role in Listeria’s virulence. The acidic part of LntA is composed of aspartic acid (<scene name='86/868192/Acidic/1'>17,8%</scene>) and the basic part is composed of lysine and arginine (<scene name='86/868192/Basic/1'>18,6%</scene>). LntA folds in a compact helical structure and is composed of 5 alpha-helix, three of them are long antiparallel helix and can be seen as the core of the protein. The two remaining helix stick out the core. The 3 first helix are named <scene name='86/868192/Helix_h1/1'>H1</scene>, <scene name='86/868192/Helix_h2/1'>H2</scene> and <scene name='86/868192/Helix_h3/1'>H3</scene>. The 2 others are <scene name='86/868192/Helix_h4ter/1'>H4</scene> and <scene name='86/868192/Helix_h5/4'>H5</scene>. These residues located in <scene name='86/868192/These_two_helix/1'>these two helixes</scene> display high RMSD values meaning that this region is likely to oscillate. In fact, by studying this protein on pymol we can see it by the thickness and redness of these helixes which means that they have a high RMSD value : | ||
| + | |||
[[Image:pymol1.jpg]] | [[Image:pymol1.jpg]] | ||
| + | |||
The flexibility of <scene name='86/868192/Helix_h4ter/1'>H4</scene> and <scene name='86/868192/Helix_h5/4'>H5</scene> may have a role in the binding to BADH1. Indeed, the positioning of this "elbow" relatively to the "trunk" formed by the less mobile regions <scene name='86/868192/Helix_h1/1'>H1</scene>, <scene name='86/868192/Helix_h2/1'>H2</scene> and <scene name='86/868192/Helix_h3/1'>H3</scene> is determining in the ligands recognition. Many amino acids may be involved in the interaction of LntA with its ligand, such as BAHD1. A <scene name='86/868192/Dilysine/1'>dilysine motif located in the elbow region of lntA at position 180/181 on the H5 helix</scene> has proven to be essential for the interaction with the transcription factor BAHD1 thanks to a conformational change <scene name='86/868192/The_lysine_180_and_181/1'>(global overview of this dilysine motif)</scene>. Indeed, when this motif is substituted by two aspartic acid amino acids (K180D/K181D by mutation of LntA), a local redistribution of the charges is observed and lntA is not able anymore to interact with BAHD1. <ref> Lebreton A, Job V, Ragon M, Le Monnier A, Dessen A, Cossart P, Bierne H. 2014. Structural basis for the inhibition of the chromatin repressor BAHD1 by the bacterial nucleomodulin LntA </ref> | The flexibility of <scene name='86/868192/Helix_h4ter/1'>H4</scene> and <scene name='86/868192/Helix_h5/4'>H5</scene> may have a role in the binding to BADH1. Indeed, the positioning of this "elbow" relatively to the "trunk" formed by the less mobile regions <scene name='86/868192/Helix_h1/1'>H1</scene>, <scene name='86/868192/Helix_h2/1'>H2</scene> and <scene name='86/868192/Helix_h3/1'>H3</scene> is determining in the ligands recognition. Many amino acids may be involved in the interaction of LntA with its ligand, such as BAHD1. A <scene name='86/868192/Dilysine/1'>dilysine motif located in the elbow region of lntA at position 180/181 on the H5 helix</scene> has proven to be essential for the interaction with the transcription factor BAHD1 thanks to a conformational change <scene name='86/868192/The_lysine_180_and_181/1'>(global overview of this dilysine motif)</scene>. Indeed, when this motif is substituted by two aspartic acid amino acids (K180D/K181D by mutation of LntA), a local redistribution of the charges is observed and lntA is not able anymore to interact with BAHD1. <ref> Lebreton A, Job V, Ragon M, Le Monnier A, Dessen A, Cossart P, Bierne H. 2014. Structural basis for the inhibition of the chromatin repressor BAHD1 by the bacterial nucleomodulin LntA </ref> | ||
Current revision
| |||||||||||
References
- ↑ ROHDE JOHN R. Listeria unwinds host’s DNA. SCIENCE, 2011 : 1271-1272
- ↑ Winter SE, Thiennimitr P et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010
- ↑ Dewoody, R., Merritt, P.M., Houppert, A.S. and Marketon, M.M. (2011), YopK regulates the Yersinia pestis type III secretion system from within host cells. Molecular Microbiology, 79: 1445-1461. https://doi.org/10.1111/j.1365-2958.2011.07534.x
- ↑ Lebreton A, Job V, Ragon M, Le Monnier A, Dessen A, Cossart P, Bierne H. 2014. Structural basis for the inhibition of the chromatin repressor BAHD1 by the bacterial nucleomodulin LntA
- ↑ Alice Lebreton. Régulations post-transcriptionnelles de l’expression génique de la cellule hôte en réponse à l’infection bactérienne. Sciences du Vivant, 2015