This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 1718
From Proteopedia
(Difference between revisions)
| (35 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | ==Human Itch G-Coupled Protein Receptors== | |
| - | == | + | <StructureSection load='7s8l' size='350' side='right' caption='Cryo-EM structure of Gq coupled MRGPRX2.' scene='90/904324/Mrgprx2/2'> |
| - | <StructureSection load=' | + | |
| - | + | ||
| - | + | ||
| - | == | + | =Introduction= |
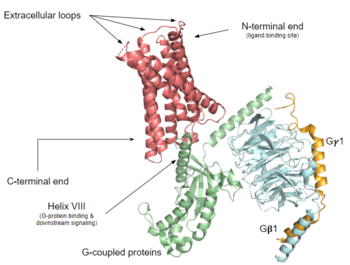
| + | [https://proteopedia.org/wiki/index.php/G_protein-coupled_receptors G protein-coupled receptors] (GPCRs) are the largest class of integral membrane proteins.<ref name="Zhang 2015">DOI 10.14348/molcells.2015.0263</ref> GPCRs<ref name= "Zhang 2015"/><ref>PMID: 20019124</ref> are divided into five families; the [https://proteopedia.org/wiki/index.php/Sandbox_Reserved_895 rhodopsin family (class A)], the [https://proteopedia.org/wiki/index.php/4ers secretin family (class B)], the [https://proteopedia.org/wiki/index.php/6wiv glutamate family (class C)], the [https://proteopedia.org/wiki/index.php/6bd4 frizzled/taste family (class F)], and the [https://en.wikipedia.org/wiki/Adhesion_G_protein-coupled_receptor adhesion family].<ref name= "Zhang 2015"/><ref name= "Zhang 2006"/> All GPCRs contain a similar seven α-helical transmembrane domain <scene name='72/727091/Full_Structure_with_Labels/1'>(TMD)</scene> that undergoes a conformation change once bound to its ligand. This conformational change then transduces a signal to a coupled, heterotrimeric G protein which then dictates whether an intracellular signaling pathway will be initiated or inhibited. The initiation of the intracellular signaling pathway occurs in response to a variety of stimuli such as light, Ca<sup>2+</sup>, peptides, different proteins, and many more stimuli. Ultimately, intracellular signaling [https://en.wikipedia.org/wiki/G_protein%E2%80%93coupled_receptor#Physiological_roles accomplishes many interesting physiological roles].<ref name= "Zhang 2015"/><ref name= "Zhang 2006">DOI 10.1371/journal.pcbi.0020013</ref> | ||
| - | == | + | Human Itch G-coupled protein receptors (GPCRs), or Mast cell-related GPCRs (MRGPRX), have been identified as pruritogenic receptors and are found in human sensory neurons, specifically in the connective tissue mast cells and dorsal root ganglia in humans.<ref name= "davidson2011">DOI: 10.1016/j.tins.2010.09.002</ref> They are classified as class A GPCRs, however, MRGPRX receptors respond to a diverse number of agonists, antagonists, and inverse agonists some of which are not typical ligands of class A receptors. MRGPRX are involved in host defense, pseudo-allergic reactions, non-histaminergic itch, periodontitis, neurogenic inflammation, and inflammatory pain.<ref name= "davidson2011"/> |
| - | + | The determination of the first structures of a ligand-activated GPCR was achieved by Robert J. Lefkowitz and Brian K. Kobilka which won them the 2012 Nobel Prize in Chemistry. They also successfully captured images of the first activated GPCR in a complex with a G protein. See [https://proteopedia.org/wiki/index.php/Nobel_Prizes_for_3D_Molecular_Structure Nobel Prizes for 3D Molecular Structure]. | |
| - | == Structural | + | ==Related Enzymes== |
| + | Among Human Itch GPCRs, <scene name='90/904324/Mrgprx2_receptor/4'>MRGPRX2</scene> is a class A GPCR that regulates mast cell degranulation and itch-related hypersensitivity reactions.<ref name="Can">DOI: 10.1038/s41586-021-04126-6</ref><ref name="Yang"/> MRGPRX2 is also a target of morphinan alkaloids, like morphine, codeine, and dextromethorphan.<ref name="Can"/><ref name="Yang"/> MRGPRX2 couples to nearly all G-protein families and subtypes with robust coupling to G<sub>q</sub> and G<sub>i</sub> families.<ref name="Can"/><ref name="Yang"/> | ||
| + | |||
| + | <scene name='90/904324/7s8p_mx4/2'>MRGPRX4</scene> is another sub-group of the MRGPRX family, which mediates cholestatic itch and is a target of nateglinide drugs.<ref name="Can"/><ref name="Yang"/> MRGPRX4 also couples to G<sub>q</sub> and G<sub>i</sub> similar to MRGPRX2. | ||
| + | |||
| + | =Structure Overview= | ||
| + | The <scene name='90/904324/Mrgprx2_receptor/4'>MRGPRX2 receptor</scene> contains 7 transmembrane helices (TM 1-7), 3 intracellular loops (ICL 1-3), and 3 extracellular loops (ECL 1-3).<ref name="Can"/> Ligand binding occurs at the N-terminus in the extracellular domain and is composed of two binding sub-pockets.<ref name="Can"/> The intracellular domain consists of helix VII and a C-terminal sequence, which binds the G-protein and promotes downstream signaling.<ref name="Can"/> | ||
| + | |||
| + | <jmol> | ||
| + | <jmolButton> | ||
| + | <script>moveto 1.0 { -413 267 871 177.06} 241.0 21.86 49.71 {118.5989314661242 127.45691253324948 105.12445970896549} 76.06737163008405 {0 0 0} 0 0 0 3.0 0.0 0.0 </script> <text>🔎Transmembrane Domain</text> | ||
| + | </jmolButton> | ||
| + | </jmol> | ||
| + | |||
| + | [[Image:Structure_overview.png|350px|center|thumb|Structural overview of MRGPRX2]] | ||
| + | |||
| + | == G-proteins and Signaling == | ||
| + | [[GTP-binding protein| G-proteins]], when paired with a GPCR, assist in signal transduction.<ref name="edward">DOI: 10.1002/pro.3526</ref><ref name="nelson">Nelson, David L. (David Lee), 1942-. (2005). Lehninger principles of biochemistry. New York :W.H. Freeman</ref> G-proteins are heterotrimeric GTPases composed of three subunits: <scene name='90/904324/Heterotrimeric_labeled/2'>α, β, and γ</scene>. The α subunit acts as the main signal mediator and contains a binding site for GDP or GTP, which acts as a biological “switch” to regulate the transmission of a signal from the activated receptor.<ref name="edward"/><ref name="nelson"/> The α subunit will dissociate and can then move in the plane of the membrane from the receptor to bind to downstream effectors to continue signal transmission and ultimately produce a cellular response.<ref name="edward"/><ref name="nelson"/> | ||
| + | |||
| + | === G<sub>q</sub> and G<sub>i</sub> Family α Subunits === | ||
| + | The actions that G-proteins induce can be classified based on the sequence homology of α subunit (G<sub>α</sub>) present.<ref name="edward"/><ref name="Kamato"/> The most well-known are referred to as G<sub>i</sub>, G<sub>s</sub>, and G<sub>q</sub>. The G<sub>s</sub> and G<sub>q</sub> proteins are stimulatory, while the G<sub>i</sub> protein is inhibitory.<ref name="edward"/><ref name="Kamato"/> In addition, proteins are classified based on the signaling pathway that they regulate. For example, G<sub>q</sub> proteins are seen in a signaling pathway that relies on phospholipase C enzymes, while G<sub>s</sub> and G<sub>i</sub> proteins are regulators of adenylate cyclase.<ref name="Kamato"/> On this page, we will be focusing solely on the structures of the G<sub>q</sub> and G<sub>i</sub> proteins and their interactions with mast-cell receptors. | ||
| + | |||
| + | G<sub>αq</sub> and G<sub>αi</sub> are proteins comprised of 359 amino acid residues, with varying sequences, that both contain a helical domain and a GTPase binding domain.<ref name= "Kamato">DOI: 10.3389/fcvm.2015.00014</ref> The GTPase binding domain is responsible for the hydrolysis of GTP as well as the binding of the β and γ subunits that form the trimeric protein structure. The helical domain contains six α helices, which are responsible for the binding of the G-protein to the coupled receptor.<ref name="Kamato"/> | ||
| + | |||
| + | The conformations of the G-proteins vary based on their association with a particular membrane receptor due to interactions between the amino acids in the N-terminus of the α subunit and the C-terminus of the receptor.<ref name="Kamato"/> | ||
| + | |||
| + | == Ligand Binding Site == | ||
| + | preface | ||
| + | <scene name='90/904324/Active_site_residues/3'>Sub-pocket 1</scene> | ||
| + | |||
| + | <jmol> | ||
| + | <jmolButton> | ||
| + | <script>moveto 1.0 { -90 -195 977 161.32} 305.32 0.0 0.0 {120.12435037299738 118.31838742815191 99.66549773755663} 68.32091981812115 {0 0 0} 0 0 0 3.0 0.0 0.0 </script> <text>🔎Active site</text> | ||
| + | </jmolButton> | ||
| + | </jmol> | ||
| + | |||
| + | ==== Sub-pocket 1 ==== | ||
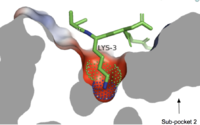
| + | [[Image:Subpocket1.jpg.png|200px|right|thumb|Cross-sectional view of electrostatic surface of MRPRX2 sub-pocket 1 interaction with lysine 3 of cortistatin-1]] | ||
| + | Sub-pocket 1 is formed by TM3, TM6, and ECL2.<ref name="Can"/> This sub-pocket is both small and deep which results in the binding of only a single amino acid residue, namely arginine or sometimes lysine.<ref name="Can"/> The binding is mediated by two key residues on the MRGPRX2 protein within the binding site: Glu164 and Asp184.<ref name="Can"/> The strong charge interactions of these two residues create a highly negatively charged electrostatic interaction within this sub-pocket.<ref name="Can"/> | ||
| + | |||
| + | ==== Sub-pocket 2 ==== | ||
| + | Sub pocket 2 is formed by TM1, TM2, TM6, and TM7.<ref name="Can"/> This binding sub-pocket is much more shallow and allows for the binding of larger structures.<ref name="Can"/> The key residues involved are Trp243 and Phe170 which allow for binding through hydrophobic interactions.<ref name="Can"/> The hydrophobicity of this binding pocket accounts for the large electrostatic difference observed between the two sub pockets. | ||
| + | |||
| + | == Differences To Most Class A GPCRs == | ||
| + | [[Image:MX2_binding_depth.png|200px|left|thumb|A. MRGPRX2 (7S8P) depicting the depth in which the ligand interacts within the receptor]][[Image:5HT2A_binding_depth.png|200px|left|thumb|B. 5HT2A (6WHA) - class A GPCR]] | ||
| + | |||
| + | The unique characteristics of <scene name='90/904324/Mrgprx2/2'>MRGPRX2</scene> in comparison to class A GPCRs provides an explanation for the differences in ligand interactions. These differences in intermolecular interactions and structural motifs contribute to the surface level ligand binding in MRGPRX2, whereas the typical ligand interaction occurs deep within the helices in class A GPCRs | ||
| + | |||
| + | ===Toggle Switch=== | ||
| + | |||
| + | The toggle switch of class A GPCRs enables the receptor to initiate the signaling cascade. However, MRGPRX2 does not contain the conserved <scene name='90/904324/Activation_mechanism/5'>toggle switch</scene> Trp. Instead, it is replaced by Gly.<ref name="Can"/> Therefore, the main residues of this motif in MRGPRX2 are Gly236, Tyr113, Phe239, and Trp243.<ref name="Can"/> As a result, TM6 is shifted closer to TM3 on the extracellular side of the membrane. This conformational change may account for the lack of ligand binding of MRGPRX2 as compared to family A receptors.<ref name="Can"/> This toggle switch swap also means that ligands, such as (R)-zinc-3573 and Cortistatin-14, bind in a different spot than ligands do on other class A GPCRs. In MRGPRX2, Gly236 is located closer to the bottom of the interface, which is the same in MRGPRX4 (Gly229). Compared with other structures, such as [https://www.rcsb.org/structure/6WHA 5-HT<sub>2A</sub>R], [https://www.rcsb.org/structure/5G53 A<sub>2A</sub>R], and [https://www.rcsb.org/structure/3SN6 β<sub>2</sub>AR], the TM6 helices of MRGPRX2 and MRGPRX4 are closer to the TM3 helix which makes the binding pocket more occluded than seen in canonical structures.<ref name="Can"/> | ||
| + | |||
| + | ===PIF/LLF motif=== | ||
| + | |||
| + | The MRGPRX2 structure does not contain the conserved <scene name='90/904324/Pifllf_motif/3'>PIF motif</scene> at the TM3-TM6 interface.<ref name="Can"/> Canonically, the PIF motif consists of a Pro, Ile, and Phe which transduce the signal produce by ligand binding through the TMD within conserved distances (5.50Å, 3.40Å, and 6.44Å respectively).<ref name="Can"/><ref>DOI: 10.1038/s41467-017-02257-x</ref> In this motif, the residues are not conserved at specific positions in the amino acid sequence but instead are conserved at distances that allow them to interact.<ref name="Can"/> | ||
| + | |||
| + | In MRGPRX2, the residues that make up TM5 have shifted down two residues making Leu194 analogous to the position of the Pro in other GPCRs. However, in MRGPRX2, Leu194 is slightly closer to the other residues in the motif at 5.48Å.<ref name="Can"/> The residue at a distance of 5.50Å in MRGPRX2 is Met196. It does not interact with the motif because it is angled away from the TM3 and TM6 interface.<ref name="Can"/> Leu194 interacts with two other residues, Leu117 and Phe232. The additional change from Ileto Leu is why the motif in MRGPRX2 is called the LLF motif.<ref name="Can"/> Compared with other structures, such as [https://www.rcsb.org/structure/6WHA 5-HT<sub>2A</sub>R], [https://www.rcsb.org/structure/5G53 A<sub>2A</sub>R], and [https://www.rcsb.org/structure/3SN6 β<sub>2</sub>AR], the TM6 helices of MRGPRX2 are closer to the TM3 helix due to the shift in residues which makes the binding pocket more occluded than seen in canonical structures.<ref name="Can"/> | ||
| + | |||
| + | ===DRY/ERC motif=== | ||
| + | |||
| + | A <scene name='Beta-2_Adrenergic_Receptor/Dry/1'>conserved DRY motif</scene> (residues 130-132) is present in all GPCRs, while the majority of class A GPCRs have a [https://proteopedia.org/wiki/index.php/A_Physical_Model_of_the_%CE%B22-Adrenergic_Receptor#conserved%20DRY%20motif conserved E/DRY motif]. However, MRGPRX2 contains an <scene name='90/904324/Erc_motif/2'>ERC motif</scene>, which replaces Tyr with Cys.<ref name="Can"/> This replacement alters the spatial organization of the helices due to the fact that Tyr is no longer present to push the helices outward.<ref name="Can"/> As a result, the binding site in MRGPRX2 is shallower which lead to surface-level ligand interactions.<ref name="Can"/> | ||
| + | |||
| + | ===Disulfide bonds=== | ||
| + | In general, class A GPCRs have a conserved disulfide bond between TM3 and ECL2.<ref name="Can"/> In contrast, the <scene name='90/904324/Mrgprx2_disulfide_bonds/3'>MRGPRX2 disulfide bond</scene> is located between Cys168 of TM4 and Cys180 of TM5, which structurally flips ECL2 to the top of TM4 and TM5.<ref name="Can"/> This creates the wide ligand-binding surface of MRGPRX2 that contributes to surface level binding which allows diverse ligand interactions.<ref name="Can"/> | ||
| + | |||
| + | ===Sodium binding site=== | ||
| + | The <scene name='90/904324/Mrgprx2_sodium_site/1'>sodium binding site</scene> of MRGPRX2 contains the conserved Asp75, however, the conserved Ser116 on TM3 seen in the [https://proteopedia.org/wiki/index.php/Neurotensin_receptor#sodium%20binding%20pocket sodium binding site] of class A GPCRs is replaced by Gly116.<ref name="Can"/> The presence of Gly as opposed to the polar Ser contributes to a lack of polar character in addition to decreasing the size of the binding pocket. The binding site generally serves as an allosteric binding site for sodium. Both factors thereby limit the binding of sodium ions to the sodium binding site in MRGPRX2. | ||
| + | |||
| + | =Mechanism= | ||
| + | Binding to the extracellular N-terminus domain triggers a transmembrane conformation change of <scene name='90/904324/Mrgprx2_receptor/4'>MRGPRX2</scene>, which demonstrates a less significant change when compared to other class A GPCRs due to the surface level binding of the ligand to MRGPRX2.<ref name="Can"/> Once ligand binding and the conformational change to the active state have taken place, the signal is relayed to the α-subunit of the heterotrimeric G-protein.<ref name="nelson"/> The α-subunit will then exchange a GDP for GTP to initiate the dissociation of the α, β, and γ subunits.<ref name="nelson"/> During this dissociation, the α-subunit is able to travel away from the receptor in the plane of the membrane to bind to downstream effectors to produce a cellular response.<ref name="nelson"/> | ||
| + | |||
| + | == Ligand interactions == | ||
| + | *<scene name='90/904324/C4880/3'>C48/80</scene> | ||
| + | **C48/80 is a peptide agonist that can bind to MRGPRX2 when it is associated with a G<sub>i</sub> or G<sub>q</sub> protein.<ref name= "Yang">DOI: 10.1038/s41586-021-04077-y</ref> The binding interactions of C48/80 with MRGPRX2 do not change whether the GPCR is associated with G<sub>i</sub> and G<sub>q</sub>, but the binding affinity of C48/80 to MRGPRX2 is different.<ref name="Yang"/> The structure of the ligand itself consists of three phenethylamine groups that are arranged in a Y shape with a semicircular arrangement.<ref name="Yang"/> Upon its binding, the Asp184 and Glu164 within sub-pocket 1 interact only with the central phenethylamine ring, forming hydrogen bonds and charge-charge interactions.<ref name="Yang"/> The central phenethylamine ring is inserted into the binding pocket at a depth of 5.6Å. This binding depth means that it is further from the toggle switch (17.7Å) than structures such as Cortistatin-14 (13.7Å).<ref name="Yang"/> The binding of this ligand induces a conformational change that causes the separation of all of the ECLs from the receptor's N-terminus.<ref name="Yang"/> | ||
| + | |||
| + | *<scene name='90/904324/Rzinc3573/4'>(R)-zinc-3573</scene> | ||
| + | **(R)-zinc-3573 is an agonist that binds to MRGPRX2 when it is associated with a G<sub>i</sub> or G<sub>q</sub> protein.<ref name="Can">DOI: 10.1038/s41586-021-04126-6</ref> This agonist is a small cationic molecule that forms largely ionic interactions with the negatively-charged sub-pocket 1 and has no interactions with sub-pocket 2.<ref name="Can"/> (R)-zinc-3573 forms hydrogen bonds and hydrophobic interactions with Asp184 and Glu164 of sub-pocket 1.<ref name="Can"/> (R)-zinc-3573 was identified through chemical screening as a MRGPRX2-selective agonist and was used to study MRGPRX2-mediated mast cell proliferation, MRGPRX2 receptor expression, mediator release and inhibition, and signaling.<ref>DOI: 10.1038/nchembio.2334</ref> | ||
| + | |||
| + | *<scene name='90/904324/Cortistatin-14/4'>Cortistatin-14</scene> | ||
| + | **Cortistatin-14 is an endogenous, cyclic, neuropeptide agonist which interacts with MRGPRX2 in the same way whether it is coupled to G<sub>i</sub> or G<sub>q</sub> proteins.<ref name="Can"/><ref name= "Jiang">DOI: 10.3389/fphar.2018.00767</ref> Cortistation-14 is widely available in many systems throughout the body and naturally functions to regulate many physiological and pathological mechanisms. These mechanisms include, but are not limited to reducing locomotion, inducing sleep, inhibiting neuronal activity, and inhibiting cell proliferation.<ref name= "Jiang"/> Cortistatin-14 is a fairly large ligand and it binds near the ECLs in subpocket-1.<ref name="Can"/> Specifically, a lysine residue (Lys3) on Cortistatin-14 binds in the negatively-charged sub-pocket 1 and forms strong charge interactions with Asp184 and Glu164.<ref name="Can"/> The remaining residues of Cortistatin-14 will extend over to sub-pocket 2 and bind through hydrophobic interactions.<ref name="Can"/> | ||
| + | |||
| + | = Clinical Relevance = | ||
| + | <scene name='90/904324/Mrgprx2_receptor/4'>MRGPRX2</scene> initiates IgE-mediated anaphylactic reactions.<ref name="porebski">DOI: 10.3389/fimmu.2018.03027</ref> MRGPRX2-mediated anaphylactic responses occur more quickly than IgE-mediated responses, but the responses also tended to be more transient.<ref name="porebski"/> Common commercial drugs, like icatibant and cetrorelix, as well as neuromuscular blocking agents activate mast cells through the MRGPRX2 pathway. | ||
| + | |||
| + | Many mutations also affect the actions of MRGPRX2. For example, a single residue mutation in sub-pocket 1 (Glu164Arg) prevented interactions between the receptor and ligands like C48/80.<ref name="porebski"/> In addition, single nucleotide polymorphisms (SNPs) have been linked to many variations of MRGPRX2 which predispose patients to hyperactivation of the receptors. Two of the most common SNPs are Asn62Thr which affects the cytoplasmic domain and Asn16His which affects the extracellular domain.<ref name="porebski"/> These mutations have been theorized to potentially protect patients from drug-induced mast cell degranulation and hypersensitivity reactions. | ||
| + | |||
| + | Interestingly, within the MRGPRX2-mediated pathway, reaction frequencies differ. In the same patients, anaphylactic reactions can occur by both IgE-mediated and MRGPRX2-mediated pathways. The presence of both mechanisms within a patient may be responsible for cross-reactivity between drugs.<ref name="porebski"/> This has led to a hypothesis that certain drugs may interact with MRGPRX2 on different active sites within the receptor. In addition, studies have also led to hypotheses that the intracellular signaling pathways triggered by the binding of varying drugs may have different intracellular responses based on the site of binding. | ||
| + | |||
| + | = See Also = | ||
| + | *[[GTP-binding protein| G proteins]] | ||
| + | *[[GTP-binding protein]] | ||
| + | *[[Pharmaceutical Drugs]] | ||
| + | *[[Nobel Prizes for 3D Molecular Structure]] | ||
| + | *[[Sandbox Reserved 895| Rhodopsin Family GPCRs]] | ||
| + | *[[4ers| Secretin Family GPCRs]] | ||
| + | *[[6wiv| Glutamate Family GPCRs]] | ||
| + | *[[6bd4| Frizzled/Taste Family GPCRs]] | ||
| + | *[https://en.wikipedia.org/wiki/Adhesion_G_protein-coupled_receptor Adhesion Family GPCRs] | ||
| - | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
| - | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | |||
| + | </StructureSection> | ||
| + | |||
| + | |||
| + | == Student contributors == | ||
| + | Madeline Beck | ||
| + | Joey Gareis | ||
Current revision
Human Itch G-Coupled Protein Receptors
| |||||||||||
Student contributors
Madeline Beck Joey Gareis