This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
SpikeProteinStructureHZ
From Proteopedia
(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 7: | Line 7: | ||
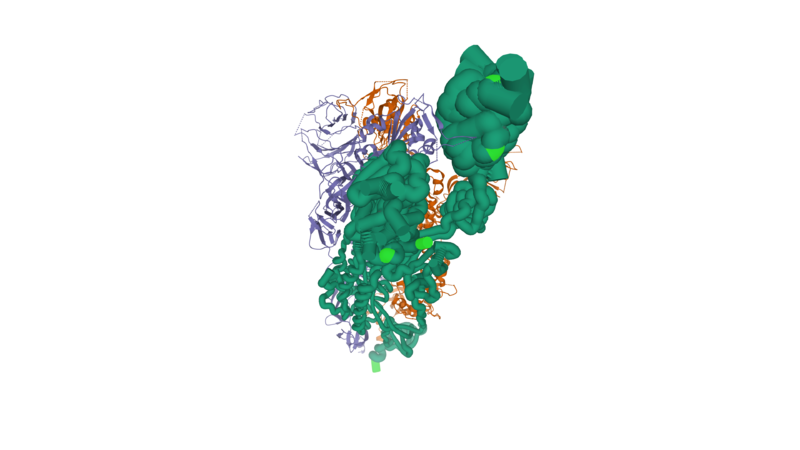
<scene name='90/903492/Ov_new_mutations/1'>Mutations @214 @371 @505 @764 were highlighted</scene>. | <scene name='90/903492/Ov_new_mutations/1'>Mutations @214 @371 @505 @764 were highlighted</scene>. | ||
Click the green text to bring up the fine structure: the newly acquired mutations, are located in the "interfacing" zone: upon cell entry, spots viral particle motifs contact ACE2. | Click the green text to bring up the fine structure: the newly acquired mutations, are located in the "interfacing" zone: upon cell entry, spots viral particle motifs contact ACE2. | ||
| + | |||
| + | We also provide another enhanced structural illustration below: | ||
| + | |||
| + | [[Image:SPG_Substructure_6VSB_new_mutations_Small.png]] | ||
| + | |||
| + | Full resolution can be found here | ||
| + | https://proteopedia.org/wiki/index.php/Image:SPG_Substructure_6VSB_new_mutations.png | ||
| + | |||
== Function == | == Function == | ||
Cell entry: Here is a simple demonstration that the newly acquired mutations in Omicron variant (OV) are located in the "interfacing" zone. | Cell entry: Here is a simple demonstration that the newly acquired mutations in Omicron variant (OV) are located in the "interfacing" zone. | ||
Current revision
Sars-CoV-2 Spike Protein Structure 02.02.2022 (Hong Y. Zhai)
| |||||||||||
You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644