We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:R. Jeremy Johnson/VKOR
From Proteopedia
< User:R. Jeremy Johnson(Difference between revisions)
(New page: ==Vitamin K Epoxide Reductase== <StructureSection load='1stp' size='340' side='right' caption='Overall Structure of Vitamin K Epoxide Reductase' scene='90/904329/Vkoroverallblue/6'> == In...) |
|||
| (4 intermediate revisions not shown.) | |||
| Line 3: | Line 3: | ||
== Introduction == | == Introduction == | ||
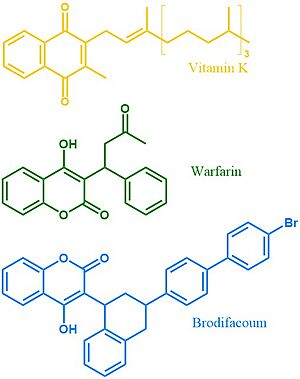
| - | Vitamin K | + | <scene name='90/904314/Vkor_structure/1'>Vitamin K Epoxide Reductase</scene> (VKOR) is a reducing enzyme composed of 4-helices that spans the endoplasmic reticulum as a transmembrane protein<ref>DOI 10.1126/science.abc5667</ref>. Its enzymatic role is reducing <scene name='90/904314/Vkor_with_ko/7'>vitamin K epoxide</scene> (KO) to Vitamin K Hydroquinone (KH2)<ref>DOI 10.1021/bi700527j</ref> (Figure 1). One of VKOR's primary roles is to assist in blood coagulation through this KH2 regeneration mechanism. |
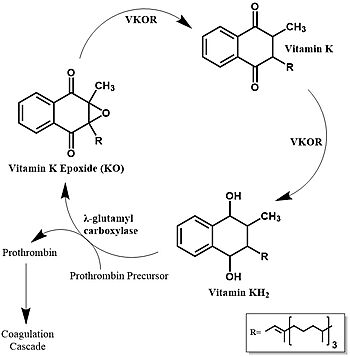
[[Image:Vitamin_k_cycle_5.jpg|350 px|left|thumb|'''Figure 1. Vitamin K Cycle''' The three states of Vitamin K are shown along with important enzymes, including VKOR. ]] | [[Image:Vitamin_k_cycle_5.jpg|350 px|left|thumb|'''Figure 1. Vitamin K Cycle''' The three states of Vitamin K are shown along with important enzymes, including VKOR. ]] | ||
| - | + | [https://en.wikipedia.org/wiki/Vitamin_K Vitamin K] is essential for [https://en.wikipedia.org/wiki/Coagulation#Coagulation_cascade blood clotting] in the body and is recycled by means of the Vitamin K Cycle <ref name="Stafford">PMID:16102054</ref> (Figure 1). The fully reduced form, KH2, is used by [https://en.wikipedia.org/wiki/Gamma-glutamyl_carboxylase#:~:text=Gamma-glutamyl%20carboxylase%20is%20an%20enzyme%20that%20catalyzes%20the,of%20the%20encoded%20enzyme%20is%20essential%20for%20hemostasis gamma-glutamyl carboxylase] to carboxylate protein-bound glutamate residues in blood clotting cofactor precursors <ref name="Blanchard">PMID:6165889</ref>. After carboxylation, the clotting cofactors (such as [https://proteopedia.org/wiki/index.php/Thrombin prothrombin]) can bind to calcium and initiatw the coagulation cascade <ref name="Swanson">PMID:6758841</ref>. During this process, KH2 becomes oxidized to Vitamin K epoxide, or KO <ref name="Blanchard">PMID:6165889</ref>. VKOR returns the epoxide to the fully reduced form for reuse by [https://en.wikipedia.org/wiki/Gamma-glutamyl_carboxylase#:~:text=Gamma-glutamyl%20carboxylase%20is%20an%20enzyme%20that%20catalyzes%20the,of%20the%20encoded%20enzyme%20is%20essential%20for%20hemostasis gamma-glutamyl carboxylase]. This transformation happens in two steps: 1) converting the epoxide to the partially oxidized Vitamin K quinone and 2) converting the quinone to the fully reduced hydroquinone (KH2) (Figure 1) <ref name="Stafford">PMID:16102054</ref>. | |
| - | [https://en.wikipedia.org/wiki/Vitamin_K Vitamin K] is essential for [https://en.wikipedia.org/wiki/Coagulation#Coagulation_cascade blood clotting] in the body and is recycled by means of the Vitamin K Cycle <ref name="Stafford">PMID:16102054</ref> (Figure 1). The fully reduced form, KH2, is used by [https://en.wikipedia.org/wiki/Gamma-glutamyl_carboxylase#:~:text=Gamma-glutamyl%20carboxylase%20is%20an%20enzyme%20that%20catalyzes%20the,of%20the%20encoded%20enzyme%20is%20essential%20for%20hemostasis gamma-glutamyl carboxylase] to carboxylate protein-bound glutamate residues in blood clotting cofactor precursors <ref name="Blanchard">PMID:6165889</ref>. After carboxylation, the clotting cofactors (such as [https://proteopedia.org/wiki/index.php/Thrombin prothrombin]) can bind to calcium and | + | |
| + | The Vitamin K Cycle and VKOR enzyme are common drug targets for thromboembolic diseases, as coagulant factor activation promotes blood clotting, which in high amounts can be dangerous and cause thromboembolic diseases such as [https://en.wikipedia.org/wiki/Stroke stroke], [https://en.wikipedia.org/wiki/Deep_vein_thrombosis deep vein thrombosis], and/or [https://en.wikipedia.org/wiki/Pulmonary_embolism pulmonary embolism]. | ||
| + | Vitamin K Epoxide Reductase is found and primarily synthesized in the [https://en.wikipedia.org/wiki/Liver liver]. | ||
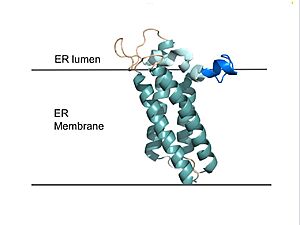
[[Image:VKOR_in_membrane.jpg|300 px|right|thumb|'''Figure 2. VKOR in Membrane''' Transmembrane helices are teal. The tan sections are the loops (loop 1, beta hairpin, cap loop, 3-4 loop). The cap helix is light blue and the anchor is dark blue.]] | [[Image:VKOR_in_membrane.jpg|300 px|right|thumb|'''Figure 2. VKOR in Membrane''' Transmembrane helices are teal. The tan sections are the loops (loop 1, beta hairpin, cap loop, 3-4 loop). The cap helix is light blue and the anchor is dark blue.]] | ||
| + | |||
| + | === Author's Note === | ||
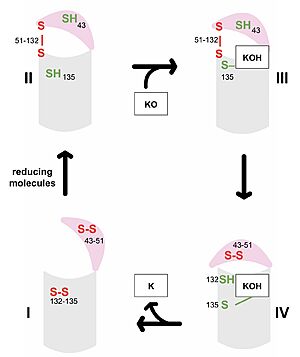
| + | To deduce VKOR structures, superfolder green flourescent protein (sGFP) was appended to the N and C termini of VKOR to provide added stability during crystallization. In some of the resolved structures, a catalytic cysteine was also mutated to serine to freeze VKOR in specific catalytic conformations. Also, due to stability issues, some experiments used human VKOR and others used VKOR from ''Takifugu rubripes''. The barrel domain has been removed from all structures used in this page and the amino acids renumbered to more closely match the published numbering in the referenced article <ref name="Liu">PMID:33154105</ref>. In the human VKOR (HsVKOR), the catalytic cysteines that play an intricate role in the reduction of Vitamin K epoxide are <scene name='90/904321/Cysteines/7'>cysteines</scene> 43, 51, 132, and 135. In ''T. rubripes'' (TrVKORL), the cysteines are 52, 55, 141, and 144. | ||
==Protein Structure== | ==Protein Structure== | ||
=== Structural Overview === | === Structural Overview === | ||
| - | VKOR consists of four <scene name='90/904330/Transmembranehelices1/5'>transmembrane helices</scene> embedded in the endoplasmic reticulum membrane (Figure 2). Helices one and two are <scene name='90/904330/Betahairpin2/5'>connected</scene> by <b><span class="text-brown">Loop 1</span></b> and the <b><span class="text-orange">beta hairpin</span></b> region which contains two of the active cysteines, Cys43 and Cys51; these cysteines, along with Cys132 and Cys135, are essential for reduction and structural changes | + | VKOR consists of four <scene name='90/904330/Transmembranehelices1/5'>transmembrane helices</scene> (<scene name='90/904321/Tm1/2'>TM1</scene>, <scene name='90/904321/Tm2/3'>TM2</scene>, <scene name='90/904321/Tm3/3'>TM3</scene>, and <scene name='90/904321/Tm4/3'>TM4 </scene>) embedded in the endoplasmic reticulum membrane (Figure 2). The transmembrane helices are located in the Endoplasmic Reticulum Luminal Region, which is the region between the ER Lumen and the [https://en.wikipedia.org/wiki/Cytosol Cytosol]. Each of these helices come together to form a central ligand binding pocket. Helices one and two are <scene name='90/904330/Betahairpin2/5'>connected</scene> by <b><span class="text-brown">Loop 1</span></b> and the <b><span class="text-orange">beta hairpin</span></b> region which contains two of the active cysteines, Cys43 and Cys51; these cysteines, along with Cys132 and Cys135, are essential for the reduction of KO and structural changes for the catalytic cycle<ref name="Liu">PMID:33154105</ref>. VKOR also has a <scene name='90/904330/Capdomain/3'>cap domain</scene> consisting of a <b><span class="text-blue">helix</span></b>, <b><span class="text-lightmagenta">loop</span></b>, and <b><span class="text-olive">anchor</span></b>. The <b><span class="text-olive">anchor</span></b> stabilizes the cap domain by attaching it to the surface of the ER membrane<ref name="Liu">PMID:33154105</ref>. The cap domain <b><span class="text-lightmagenta">loop</span></b> helps stabilize one of the substrate-binding amino acids, Asn80<ref name="Liu">PMID:33154105</ref>. The cap domain <b><span class="text-blue">helix</span></b> is involved in stabilization of certain disulfide bonds and structural changes as part of the catalytic cycle<ref name="Liu">PMID:33154105</ref>. The substrate binding pocket works in conjunction with the cap domain and the anchor to facilitate conformational transitions from the <scene name='90/904314/Open_conformation/3'>open conformation</scene> to the <scene name='90/904314/Closed_conformation/6'>closed conformation</scene> once a substrate binds. |
| - | === | + | === Cap Domain === |
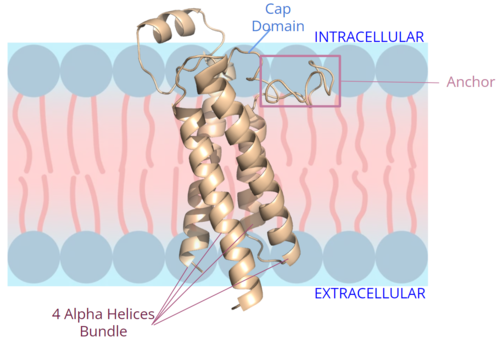
| - | + | [[Image:New membrane pic bundle.png|500 px|right|thumb|Figure 2. Orientation and interactions of the VKOR components cap domain, anchor domain, and helical bundle within the cell membrane.]] | |
| + | The VKOR <scene name='90/904314/Cap_domain/8'>cap domain</scene> is located right above the helices of VKOR on the intracellular membrane. The cap domain has a helical shape and is located in close proximity to two other domains: the Anchor domain and beta hairpin. This combination of domains help to maintain the proper orientation in the membrane. The cap domain assists with activating Vitamin K as it induces the structural change of VKOR from the open conformation to the closed conformation upon substrate binding. Cap rearrangement and transition to the closed conformation initiates a domino effect through the catalytic mechanism<ref>DOI 10.1074/jbc.RA120.015401</ref>. The cap domain has critical interactions that stabilize the closed conformation including a <scene name='90/904314/Disulfide_bridge_stabilization/8'>disulfide bridge</scene> between Cys43 and Cys51, and polar interactions from D44. These interactions are broken up by reactive cysteines to induce different conformations and help facilitate this transition from the open conformation to the closed conformation during the activation of Vitamin K. | ||
| + | |||
| + | === Anchor === | ||
| + | The <scene name='90/904314/Anchor_domain/12'>anchor domain</scene> protrudes from the side of VKOR to stabilize the enzyme within the membrane. It sits on top of the membrane and maintain proper membrane proximity for VKOR activity (Figure 2). To accomplish proper membrane orientation, hydrophilic residues on the anchor domain are positioned to interact with the outer hydrophilic leaflet of the bilipid membrane, while the hydrophobic residues on the anchor have strong interactions with the inner hydrophobic leaflet of the bilipid membrane. These <scene name='90/904314/Anchor_domain/13'>polar and nonpolar interactions</scene> control proper membrane arrangement, Vitamin K binding, and VKOR activation via cap domain movement. The anchor also stabilizes the cap domain covering the central binding pocket and keeping substrates bound in the active site. | ||
=== Active Site === | === Active Site === | ||
| Line 45: | Line 54: | ||
<references/> | <references/> | ||
==Student Contributors== | ==Student Contributors== | ||
| - | Izabella Jordan | + | Kiana Jackson |
| + | |||
| + | Izabella Jordan | ||
| + | |||
| + | George Papadeas | ||
| + | |||
| + | Bianca Pontrelli | ||
| + | |||
| + | Anna Pressel | ||
| + | |||
| + | Emma Varness | ||
Current revision
Vitamin K Epoxide Reductase
| |||||||||||
References
- ↑ Liu S, Li S, Shen G, Sukumar N, Krezel AM, Li W. Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation. Science. 2020 Nov 5. pii: science.abc5667. doi: 10.1126/science.abc5667. PMID:33154105 doi:http://dx.doi.org/10.1126/science.abc5667
- ↑ Jin DY, Tie JK, Stafford DW. The conversion of vitamin K epoxide to vitamin K quinone and vitamin K quinone to vitamin K hydroquinone uses the same active site cysteines. Biochemistry. 2007 Jun 19;46(24):7279-83. doi: 10.1021/bi700527j. Epub 2007 May, 25. PMID:17523679 doi:http://dx.doi.org/10.1021/bi700527j
- ↑ 3.0 3.1 Stafford DW. The vitamin K cycle. J Thromb Haemost. 2005 Aug;3(8):1873-8. doi: 10.1111/j.1538-7836.2005.01419.x. PMID:16102054 doi:http://dx.doi.org/10.1111/j.1538-7836.2005.01419.x
- ↑ 4.0 4.1 Blanchard RA, Furie BC, Jorgensen M, Kruger SF, Furie B. Acquired vitamin K-dependent carboxylation deficiency in liver disease. N Engl J Med. 1981 Jul 30;305(5):242-8. doi: 10.1056/NEJM198107303050502. PMID:6165889 doi:http://dx.doi.org/10.1056/NEJM198107303050502
- ↑ Swanson JC, Suttie JW. Vitamin K dependent in vitro production of prothrombin. Biochemistry. 1982 Nov 9;21(23):6011-8. doi: 10.1021/bi00266a044. PMID:6758841 doi:http://dx.doi.org/10.1021/bi00266a044
- ↑ 6.00 6.01 6.02 6.03 6.04 6.05 6.06 6.07 6.08 6.09 6.10 Liu S, Li S, Shen G, Sukumar N, Krezel AM, Li W. Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation. Science. 2020 Nov 5. pii: science.abc5667. doi: 10.1126/science.abc5667. PMID:33154105 doi:http://dx.doi.org/10.1126/science.abc5667
- ↑ Shen G, Cui W, Cao Q, Gao M, Liu H, Su G, Gross ML, Li W. The catalytic mechanism of vitamin K epoxide reduction in a cellular environment. J Biol Chem. 2021 Jan-Jun;296:100145. doi: 10.1074/jbc.RA120.015401. Epub 2020, Dec 10. PMID:33273012 doi:http://dx.doi.org/10.1074/jbc.RA120.015401
- ↑ 8.0 8.1 Patel S, Singh R, Preuss CV, Patel N. Warfarin PMID:29261922
- ↑ Wu S, Chen X, Jin DY, Stafford DW, Pedersen LG, Tie JK. Warfarin and vitamin K epoxide reductase: a molecular accounting for observed inhibition. Blood. 2018 Aug 9;132(6):647-657. doi: 10.1182/blood-2018-01-830901. Epub 2018, May 9. PMID:29743176 doi:http://dx.doi.org/10.1182/blood-2018-01-830901
- ↑ 10.0 10.1 Chong YK, Mak TW. Superwarfarin (Long-Acting Anticoagulant Rodenticides) Poisoning: from Pathophysiology to Laboratory-Guided Clinical Management. Clin Biochem Rev. 2019 Nov;40(4):175-185. doi: 10.33176/AACB-19-00029. PMID:31857739 doi:http://dx.doi.org/10.33176/AACB-19-00029
Student Contributors
Kiana Jackson
Izabella Jordan
George Papadeas
Bianca Pontrelli
Anna Pressel
Emma Varness