BASIL2022GV3R8E
From Proteopedia
(Difference between revisions)
| (4 intermediate revisions not shown.) | |||
| Line 14: | Line 14: | ||
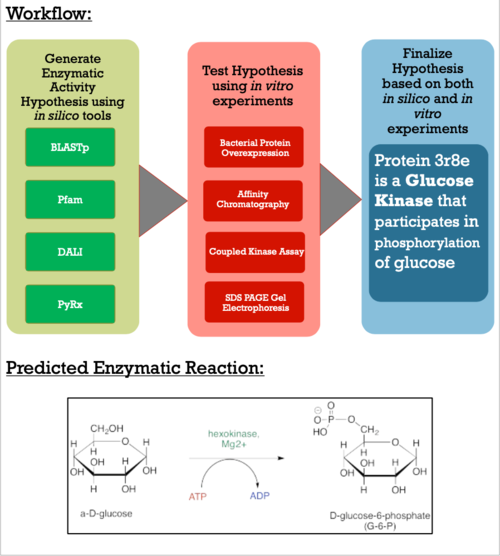
| - | From here, we were able to form the conclusion that our POI interacts with glucose based on the alignment with a known hexokinase. To validate that glucose actually binds and interacts with our protein of interest, we conducted a PyRx in silico docking experiment with a total of five hexose substrates. Other substrates tested include fructose, galactose, lactose, and ribose, however, experimental in silico docking results for those substrates were significantly less than glucose. Along with the PyRx docking, we visualized <scene name='90/904995/Glucose_and_atp/1'>ATP and glucose</scene> within the proposed active site in the PyMol visualization software tool. We also were then able to find which active site amino acid were crucial to binding, which are highlighted <scene name='90/904995/ | + | From here, we were able to form the conclusion that our POI interacts with glucose based on the alignment with a known hexokinase. To validate that glucose actually binds and interacts with our protein of interest, we conducted a PyRx in silico docking experiment with a total of five hexose substrates. Other substrates tested include fructose, galactose, lactose, and ribose, however, experimental in silico docking results for those substrates were significantly less than glucose. Along with the PyRx docking, we visualized <scene name='90/904995/Glucose_and_atp/1'>ATP and glucose</scene> within the proposed active site in the PyMol visualization software tool. We also were then able to find which active site amino acid were crucial to binding, which are highlighted <scene name='90/904995/3r8e_amino_acids_updated/3'>here</scene>. The binding affinity of glucose was -5.1 kcal/mol, which strengthens our idea that glucose is phosphorylated by our protein of interest. The confidence behind our in silico results allowed us to move into testing our hypothesis in vitro and because ATP aids in the phosphorylation of glucose, an <scene name='90/904995/3r8ec_w_glc_and_atp/1'>interactive structure</scene> has been provided to represent interactions of glucose and ATP in the active site. |
| Line 21: | Line 21: | ||
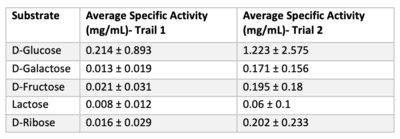
Once we felt confident enough to finalize our substrate hypothesis, we began testing in vitro. Beginning with bacterial protein overexpression and affinity chromatography, we were able to purify our POI and begin testing with real substrates. Below are the results of our Uncoupled Kinase Assay, reported in terms of specific activity (mg/mL). Our results from this assay further supports our idea of protein 3r8e assisting in the phosphorylation of glucose. A total of five hexose substrates were tested in vitro, detailed in the table below. Based on these results, we were able to strengthen our initial hypothesis and continue characterization. | Once we felt confident enough to finalize our substrate hypothesis, we began testing in vitro. Beginning with bacterial protein overexpression and affinity chromatography, we were able to purify our POI and begin testing with real substrates. Below are the results of our Uncoupled Kinase Assay, reported in terms of specific activity (mg/mL). Our results from this assay further supports our idea of protein 3r8e assisting in the phosphorylation of glucose. A total of five hexose substrates were tested in vitro, detailed in the table below. Based on these results, we were able to strengthen our initial hypothesis and continue characterization. | ||
| - | [[Image: | + | [[Image:SA_1.png|400px|]] |
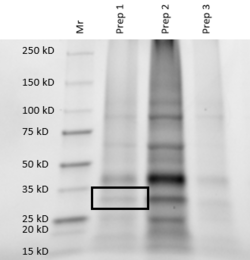
For further validation, we conducted an SDS analysis and provided below is the gel image. Indicated by the black box is our POI, around 34 kDa. Results were not as clear as anticipated, and in future studies, we would need to utilize different chromatography methods to yield higher quality protein concentrations and conduct a pre and post induction to visualize the purity of our protein. | For further validation, we conducted an SDS analysis and provided below is the gel image. Indicated by the black box is our POI, around 34 kDa. Results were not as clear as anticipated, and in future studies, we would need to utilize different chromatography methods to yield higher quality protein concentrations and conduct a pre and post induction to visualize the purity of our protein. | ||
| Line 37: | Line 37: | ||
After validating our results, we now look to take our findings to a micropublication website for undergraduate research. By doing this, not only will our work be forward facing and available to the science community, but it also allows for collaboration and further questions to be asked. After completing the micropublication, we look to continue to develop research strategies for putative kinases, as the PDB has thousands of proteins with unsolved functions. We will do this by combining machine learning, data science, and lab work to allow undergraduate students and scientist to effectively research and study putative kinase structures and functions. | After validating our results, we now look to take our findings to a micropublication website for undergraduate research. By doing this, not only will our work be forward facing and available to the science community, but it also allows for collaboration and further questions to be asked. After completing the micropublication, we look to continue to develop research strategies for putative kinases, as the PDB has thousands of proteins with unsolved functions. We will do this by combining machine learning, data science, and lab work to allow undergraduate students and scientist to effectively research and study putative kinase structures and functions. | ||
| - | + | ||
== References == | == References == | ||
<references/> | <references/> | ||
| Line 51: | Line 51: | ||
6. The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC. | 6. The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC. | ||
| + | |||
| + | [[Category: BASIL]] | ||
Current revision
Characterization of the 3r8e Protein, a Novel Glucose Kinase
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
Dalton Dencklau, Michel Evertsen, Bonnie Hall, Jaime Prilusky