We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1778
From Proteopedia
(Difference between revisions)
| (26 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
{{Template:CH462_Biochemistry_II_2023}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | {{Template:CH462_Biochemistry_II_2023}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
==Your Heading Here (maybe something like 'Structure')== | ==Your Heading Here (maybe something like 'Structure')== | ||
| - | <StructureSection load=' | + | <StructureSection load='7UPI' size='350' frame='true' |
| + | side='right' caption='SMP' scene=’’> | ||
This is a default text for your page ''''''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | This is a default text for your page ''''''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
== Function == | == Function == | ||
| + | <scene name='95/952705/Shoc2_mras_interaction/2'>hydrogen bonding</scene> | ||
| + | <scene name='95/952706/Shoc2_mras_interaction/2'>hydrogen bonding</scene> | ||
| - | == | + | <scene name='95/952705/Shoc2_pp1c_interaction/1'>PP1C binds</scene> |
| + | <scene name='95/952706/Shoc2_pp1c_interaction/3'>PP1C binds</scene> | ||
| - | == | + | <scene name='95/952705/Shoc2_rvxf/3'>PVxF motif</scene> |
| + | <scene name='95/952706/Shoc2_rvxf/1'>RVxF motif</scene> | ||
| - | == | + | <scene name='95/952705/Mras_pp1c_interaction/2'>MRAS binds to PP1C</scene> |
| + | <scene name='95/952706/Mras_pp1c_interaction/2'>MRAS binds to PP1C</scene> | ||
| - | = | + | =Structure= |
| + | ==Overview== | ||
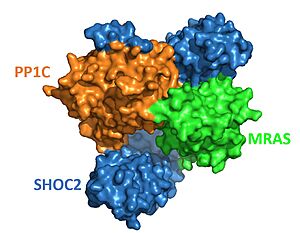
| - | + | The protein combines 3 separate '''(see notes for more)'''. SHOC2, PP1C, and MRAS, to form the active protein (SMP complex), as seen in Figure 2<Ref name='Hauseman'>Hauseman, Z.J., Fodor, M., Dhembi, A. et al. Structure of the MRAS–SHOC2–PP1C phosphatase complex. Nature 609, 416–423 (2022). doi: 10.1038/s41586-022-05086-1. [https://doi.org/10.1038/s41586-022-05086-1. DOI:10.1038/s41586-022-05086-1]. </Ref>. The SMP complex was determined via cryo-electron microscopy as well as x-ray diffraction. The overall structure has PP1C and MRAS bound within the concave surface of SHOC2, leaving the catalytic site of PP1C and the GTP binding cleft in MRAS exposed. | |
| - | + | ||
| - | = MRAS = | + | ==SHOC2== |
| + | [[Image:SHOC2-PP1C-MRAS Surface.JPG|300px|right|thumb|<font size="2"><div style="text-align: center;">'''Figure 2'''. Surface representation of SHOC2-PP1C-MRAS ([https://www.rcsb.org/structure/7UPI PDB 7upi]). SHOC2 (blue), PP1C (orange) and MRAS (green). </div></font>]] | ||
| + | SHOC2 is a scaffolding protein which acts as a cradle to bind PP1C and MRAS '''(REF)''', serving as an aggregation point for these 3 signaling proteins. <scene name='95/952705/Shoc2_structure/1'>SHOC2</scene> is a leucine rich repeat ([https://en.wikipedia.org/wiki/Leucine-rich_repeat LRR]) protein consisting of 20 consecutive <scene name='95/952706/Shoc2_structure/6'>LRR motifs</scene>. LRR motifs form an an extended '''BETA''' sheet on the inner concave surface of SHOC2 with alpha helices facing outward, '''which help bind the whole protein'''. These LRR motifs result in a largely hydrophobic core '''within the concaved region'''.<Ref name= 'Hahn'> Kwon, J. J., & Hahn, W. C. A Leucine-Rich Repeat Protein Provides a SHOC2 the RAS Circuit: a Structure-Function Perspective. Molecular and cellular biology, 41(4), e00627-20 (2021). doi:10.1128/MCB.00627-20. [http://doi.org/10.1128/MCB.00627-20. DOI: 10.1128/MCB.00627-20]. </Ref>. | ||
| - | < | + | ==PP1C== |
| - | + | <scene name='95/952705/Pp1c_structure/1'>PP1C</scene> is the catalytic domain of the phosphatase enzyme [https://www.ncbi.nlm.nih.gov/gene/5499 PP1], which removes reversible phosphorylations from signaling proteins. PP1C is a serine/threonine phosphatase that is involved in cellular signaling pathways that control cell growth, division, and metabolism '''(REF)''' '''and explain more, see notes'''. | |
| - | = PP1C = | + | ==MRAS== |
| + | <scene name='95/952705/Mras_structure/4'>MRAS</scene> is a monomeric GTPase and is anchored in the cell membrane. When MRAS binds GTP, it becomes active, and '''the rest can only bind when this is activated'''<ref name="Hauseman" />. MRAS is a subvariant of the RAS protein and therefore shares most of it's regulatory and effector interactions<Ref name= 'Young'>Young, L., Rodriguez-Viciana, P. MRAS: A Close but Understudied Member of the RAS Family. Cold Spring Harbor Perspectives in Medicine (2018). doi: 10.1101/cshperspect.a033621. [https://perspectivesinmedicine.cshlp.org/content/8/12/a033621.full.pdf+html. DOI: 0.1101/cshperspect.a033621]. </Ref>. '''While other RAS variants bind in complex with SHOC2 and PP1 to allow it to have phosphatase activity, MRAS binds the tightest.''' | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ==Stabilizing Interactions== | ||
| + | <scene name='95/952706/Shoc2_mras_interaction/2'>MRAS binds to SHOC2</scene> exclusively through its concave LRRs<Ref name='Kwan'>Kwon, J.J., Hajian, B., Bian, Y. et al. Structure–function analysis of the SHOC2–MRAS–PP1C holophosphatase complex. Nature 609, 408–415 (2022).doi: 10.1038/s41586-022-04928-2. [https://doi.org/10.1038/s41586-022-04928-2. DOI:10.1038/s41586-022-04928-2] </Ref>, primarily by the descending loop and strands of LRR domains 2-10. <scene name='95/952706/Shoc2_pp1c_interaction/3'>PP1C binds</scene> with the ascending loops of the SHOC2 LRR regions, and is further engaged through the N-terminal region of SHOC2 containing the <scene name='95/952706/Shoc2_rvxf/1'>RVxF motif</scene> <Ref name= 'Jajian'>Kwon, J., Jajian, B., Bian, Y. et al. Comprehensive structure-function evaluation of the SHOC2 holophosphatase reveals disease mechanisms and therapeutic opportunities. In: Proceedings of the American Association for Cancer Research Annual Meeting 2022. [https://aacrjournals.org/cancerres/article/82/12_Supplement/LB029/699443. DOI: 10.1158/1538-7445.AM2022-LB029]. </Ref>. The initial forming of the complex begins with SHOC2-PP1C engagement, then is completed and stabilized by the GTP-loaded MRAS binding '''(apparently described differently elsewhere?)''' <Ref name="Jajian" />. Once associated with SHOC2, <scene name='95/952706/Mras_pp1c_interaction/2'>MRAS binds to PP1C</scene>. Binding to MRAS localizes the other two proteins to the RAS signaling regions of the membrane to begin cellular signaling.<ref name="Kwan" />. | ||
| + | |||
| + | |||
| + | =Implications= | ||
| + | The SHOC2-MRAS-PP1C complex's key role in the regulation of the MAPK-RAF pathway means that minor changes in its structure or function can have drastic biological consequences. Unregulated activation of the MAPK pathway is one of the most common causes of human cancer due to unchecked cell division and proliferation '''(REF)'''. Mutations that stabilize the interactions of the SMP complex enhances PP1C phosphatase activity <Ref name="Jajian" />, leading to increased RAF signaling and accelerated cell division. | ||
| + | |||
| + | The unregulated MAPK pathway is also responsible for a multitude of developmental disorders commonly known as [https://dceg.cancer.gov/research/what-we-study/rasopathies#:~:text=RASopathies%20are%20a%20group%20of,to%20grow%20and%20work%20properly. RASopathies] <Ref name="Lavoie" />. One such RASopathy caused by the mutation of a RAF kinase is known as [https://www.mayoclinic.org/diseases-conditions/noonan-syndrome/symptoms-causes/syc-20354422 Noonan Syndrome] (NS), which enhances complex formation '''(WHY)'''. NS is a genetic disorder that can prevent normal development in different parts of the body in a variety of ways. NS patients often have unusual facial characteristics, short stature, a variety of heart defects and disease, and other physical problems and developmental delays. | ||
| + | |||
| + | === SHOC2 === | ||
| + | SHOC2 is a domain which acts as a cradle to bind PP1C and MRAS. The | ||
| + | <scene name='95/952706/Shoc2_structure/7'>structure of SHOC2</scene> | ||
| + | is a leucine rich repeat (LRR) protein that consists of 20 consecutive | ||
| + | <scene name='95/952706/Shoc2_structure/6'>LRR regions</scene> | ||
| + | . This motif results in an extended beta sheet on the inner concavity of the protein surface with alpha helices facing outward. This results in a largely hydrophobic core. | ||
| + | |||
| + | protein phosphatase 1 catalytic subunit ( <scene name='95/952706/Pp1c_structure/3'>PP1C</scene> ) is highly characterized of serine/ threonine phosphatase. PP1C | ||
| + | <scene name='95/952706/Shoc2_pp1c_interaction/2'>associates</scene>> | ||
| + | associates with the ascending loops of the SHOC2 LRR regions. | ||
| + | |||
| + | === MRAS === | ||
| + | <scene name='95/952706/Mras_structure/1'>MRAS</scene> | ||
| + | binds to SHOC2 primarily by the descending loop and strands of each LRR domains 2-10. Once associated with SHOC2, it binds to PP1C and guides the holoenzyme complex to the cell membrane to begin signaling. | ||
| + | |||
| + | === PP1C === | ||
| - | <StructureSection load='7UP1' size='350' frame='true' | ||
| - | side='right' caption='PP1C domain of SMP' scene=’’> | ||
= Interactions = | = Interactions = | ||
| + | <scene name='95/952706/Shoc2_pp1c_interaction/1'>SHOC2 and PP1C interaction</scene> | ||
| + | <scene name='95/952706/Shoc2_mras_interaction/1'>SHOC2 and MRAS interactions</scene> | ||
| + | <scene name='95/952706/Mras_pp1c_interaction/1'>MRAS and PP1C interactions</scene> | ||
This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
Current revision
| This Sandbox is Reserved from February 27 through August 31, 2023 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1765 through Sandbox Reserved 1795. |
To get started:
More help: Help:Editing |
Your Heading Here (maybe something like 'Structure')
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ 3.0 3.1 Hauseman, Z.J., Fodor, M., Dhembi, A. et al. Structure of the MRAS–SHOC2–PP1C phosphatase complex. Nature 609, 416–423 (2022). doi: 10.1038/s41586-022-05086-1. DOI:10.1038/s41586-022-05086-1.
- ↑ Kwon, J. J., & Hahn, W. C. A Leucine-Rich Repeat Protein Provides a SHOC2 the RAS Circuit: a Structure-Function Perspective. Molecular and cellular biology, 41(4), e00627-20 (2021). doi:10.1128/MCB.00627-20. DOI: 10.1128/MCB.00627-20.
- ↑ Young, L., Rodriguez-Viciana, P. MRAS: A Close but Understudied Member of the RAS Family. Cold Spring Harbor Perspectives in Medicine (2018). doi: 10.1101/cshperspect.a033621. DOI: 0.1101/cshperspect.a033621.
- ↑ 6.0 6.1 Kwon, J.J., Hajian, B., Bian, Y. et al. Structure–function analysis of the SHOC2–MRAS–PP1C holophosphatase complex. Nature 609, 408–415 (2022).doi: 10.1038/s41586-022-04928-2. DOI:10.1038/s41586-022-04928-2
- ↑ 7.0 7.1 7.2 Kwon, J., Jajian, B., Bian, Y. et al. Comprehensive structure-function evaluation of the SHOC2 holophosphatase reveals disease mechanisms and therapeutic opportunities. In: Proceedings of the American Association for Cancer Research Annual Meeting 2022. DOI: 10.1158/1538-7445.AM2022-LB029.
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedLavoie