User:Eduardo Soares/Sandbox 1
From Proteopedia
< User:Eduardo Soares(Difference between revisions)
| (4 intermediate revisions not shown.) | |||
| Line 3: | Line 3: | ||
The PDB code 2RDE represents a PilZ protein complexed with cyclic diguanylate monophosphate (c-di-GMP) in ''Vibrio cholera'' there is a molecular interaction involved in the regulation of bacterial biofilm formation and motility. The complex is formed by the binding of the small signaling molecule c-di-GMP to a protein domain known as PilZ. This interaction plays a crucial role in bacterial physiology and is implicated in various cellular processes. <ref>Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007 Dec 12;26(24):5153-66. doi: 10.1038/sj.emboj.7601918. Epub 2007 Nov 22. PMID: 18034161; PMCID: PMC2140105.</ref> | The PDB code 2RDE represents a PilZ protein complexed with cyclic diguanylate monophosphate (c-di-GMP) in ''Vibrio cholera'' there is a molecular interaction involved in the regulation of bacterial biofilm formation and motility. The complex is formed by the binding of the small signaling molecule c-di-GMP to a protein domain known as PilZ. This interaction plays a crucial role in bacterial physiology and is implicated in various cellular processes. <ref>Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007 Dec 12;26(24):5153-66. doi: 10.1038/sj.emboj.7601918. Epub 2007 Nov 22. PMID: 18034161; PMCID: PMC2140105.</ref> | ||
| - | <StructureSection load=' | + | <StructureSection load='2RDE' size='340' side='right' caption='Caption for this structure' scene=''> |
== PilZ domain == | == PilZ domain == | ||
| Line 10: | Line 10: | ||
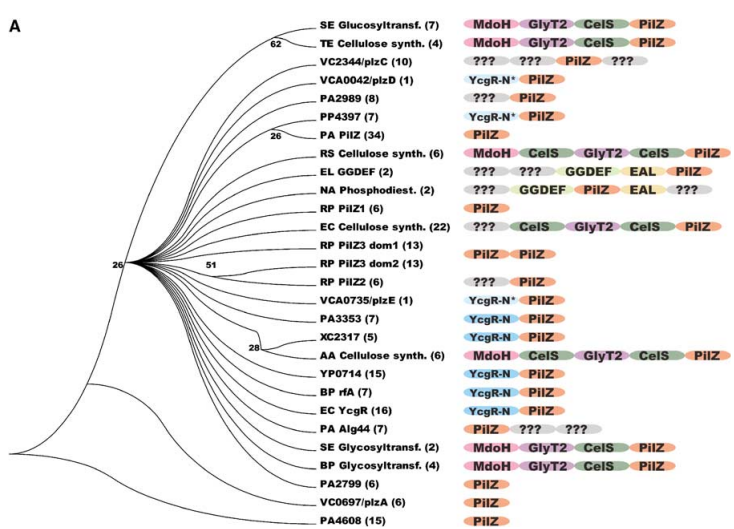
Notably, the PilZ protein in'' Pseudomonas aeruginosa'' contains a single 118-residue domain exhibiting substantial sequence similarity to the C-terminus of BcsA. Within the pil operon, responsible for the synthesis and functionality of pili, PilZ is among the few proteins with an unknown function. This operon plays a role in the transition from motile to sessile growth modes, although the precise regulatory mechanisms remain elusive. Strains lacking the PilZ domain (ΔpilZ) can produce normal levels of pilin protomers but are unable to assemble functional pili. | Notably, the PilZ protein in'' Pseudomonas aeruginosa'' contains a single 118-residue domain exhibiting substantial sequence similarity to the C-terminus of BcsA. Within the pil operon, responsible for the synthesis and functionality of pili, PilZ is among the few proteins with an unknown function. This operon plays a role in the transition from motile to sessile growth modes, although the precise regulatory mechanisms remain elusive. Strains lacking the PilZ domain (ΔpilZ) can produce normal levels of pilin protomers but are unable to assemble functional pili. | ||
| - | Furthermore, other PilZ-domain proteins have been implicated in the regulation of motility across various organisms. For instance, the YcgR protein in ''Escherichia coli'' and the DgrA and DgrB proteins in ''Caulobacter crescentus'' have demonstrated involvement in controlling motility under high cytosolic c-di-GMP concentrations. Notably, certain PilZ domains are situated at the C-terminus of proteins participating in processes sensitive to c-di-GMP levels. Alg44 from P. aeruginosa represents another example, functioning as an essential protein in the c-di-GMP-regulated process of alginate biosynthesis. These discoveries provide robust evidence supporting the hypothesis that the majority of PilZ domains function as effectors, binding c-di-GMP, and exerting a critical role in the regulation of various cellular processes. <ref> Pecina, Anna, et al. The Stand-Alone PilZ-Domain Protein MotL Specifically Regulates the Activity of the Secondary Lateral Flagellar System in Shewanella putrefaciens. Frontiers in Microbiology, vol. 12, 2021. Frontiers, https://www.frontiersin.org/articles/10.3389/fmicb.2021.668892. </ref> <ref> | + | Furthermore, other PilZ-domain proteins have been implicated in the regulation of motility across various organisms. For instance, the YcgR protein in ''Escherichia coli'' and the DgrA and DgrB proteins in ''Caulobacter crescentus'' have demonstrated involvement in controlling motility under high cytosolic c-di-GMP concentrations. Notably, certain PilZ domains are situated at the C-terminus of proteins participating in processes sensitive to c-di-GMP levels. Alg44 from P. aeruginosa represents another example, functioning as an essential protein in the c-di-GMP-regulated process of alginate biosynthesis. These discoveries provide robust evidence supporting the hypothesis that the majority of PilZ domains function as effectors, binding c-di-GMP, and exerting a critical role in the regulation of various cellular processes. <ref> Pecina, Anna, et al. The Stand-Alone PilZ-Domain Protein MotL Specifically Regulates the Activity of the Secondary Lateral Flagellar System in Shewanella putrefaciens. Frontiers in Microbiology, vol. 12, 2021. Frontiers, https://www.frontiersin.org/articles/10.3389/fmicb.2021.668892. </ref> <ref>Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007 Dec 12;26(24):5153-66. doi: 10.1038/sj.emboj.7601918. Epub 2007 Nov 22. PMID: 18034161; PMCID: PMC2140105.</ref> |
| Line 20: | Line 20: | ||
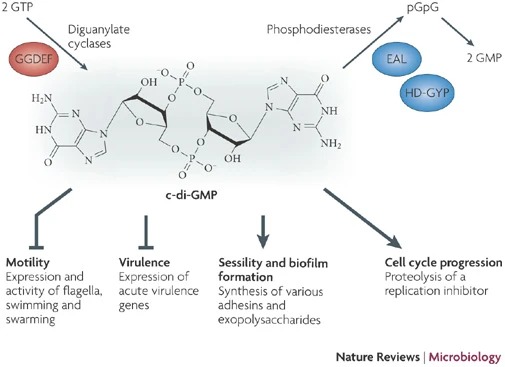
The significance of c-di-GMP regulatory systems gained considerable attention following the advent of whole genome sequencing, revealing the widespread presence of GGDEF and EAL domains in bacterial species, often with a large number of encoded proteins. Artificial modulation of cellular c-di-GMP levels through overproduction of GGDEF domain proteins led to enhanced adhesin and biofilm matrix component synthesis, while impairing motility and acute virulence functions. Conversely, overproduction of EAL domain proteins produced opposite phenotypes (Figure 1). Current research aims to assign specific molecular functions to GGDEF, EAL, and HD-GYP domain proteins and unravel their integration into complex regulatory networks. Notably, recent findings have identified biologically active GGDEF and EAL domain proteins lacking DGC or PDE activity, exemplifying their involvement in transcriptional regulation and RNA degradation. | The significance of c-di-GMP regulatory systems gained considerable attention following the advent of whole genome sequencing, revealing the widespread presence of GGDEF and EAL domains in bacterial species, often with a large number of encoded proteins. Artificial modulation of cellular c-di-GMP levels through overproduction of GGDEF domain proteins led to enhanced adhesin and biofilm matrix component synthesis, while impairing motility and acute virulence functions. Conversely, overproduction of EAL domain proteins produced opposite phenotypes (Figure 1). Current research aims to assign specific molecular functions to GGDEF, EAL, and HD-GYP domain proteins and unravel their integration into complex regulatory networks. Notably, recent findings have identified biologically active GGDEF and EAL domain proteins lacking DGC or PDE activity, exemplifying their involvement in transcriptional regulation and RNA degradation. | ||
| - | [[Image:Figura2.jpg]] <ref>Hengge, R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7, 263–273 (2009). https://doi.org/10.1038/nrmicro2109< | + | [[Image:Figura2.jpg]] <ref>Hengge, R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7, 263–273 (2009). https://doi.org/10.1038/nrmicro2109</ref> |
| - | + | ||
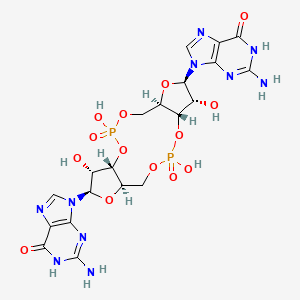
==Molecular Interaction== | ==Molecular Interaction== | ||
| Line 46: | Line 45: | ||
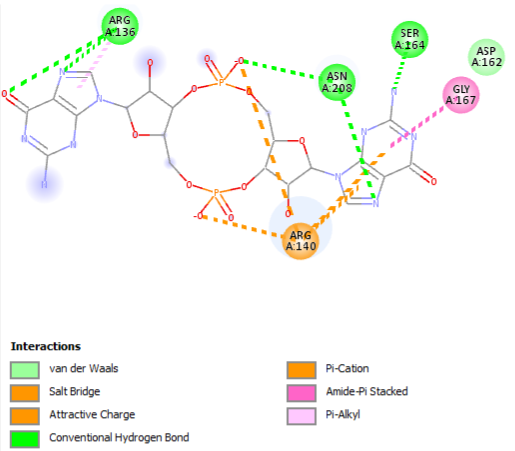
[[Image:3D sitio2.png]] | [[Image:3D sitio2.png]] | ||
| + | |||
| + | [[Image:3D sítio.png]] | ||
== Structural highlights == | == Structural highlights == | ||
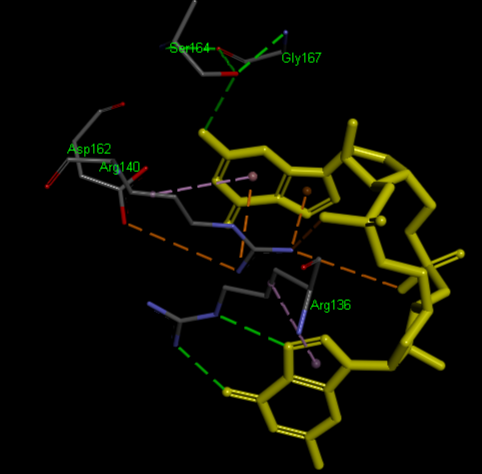
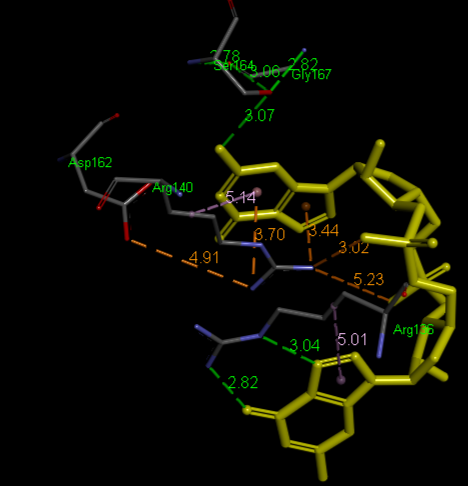
Highlight representation of the the <scene name="/12/3456/Sample/1">c-di-GMP</scene> and <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein, putting in evidence the alpha helix. | Highlight representation of the the <scene name="/12/3456/Sample/1">c-di-GMP</scene> and <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein, putting in evidence the alpha helix. | ||
| + | ===Crystal structure of VCA0042 complexed with c-di-GMP ([[2rde]])=== | ||
| + | |||
| + | '''Pilz domain'''<br /> | ||
| + | |||
| + | <scene name='2rde/Plzd_start/1'>PilZ</scene> | ||
| + | |||
| + | <scene name='2rde/Plzd_active/1'>Binding sites</scene> (R136, R140, D162, S164, G167) | ||
| + | |||
| + | <scene name='2rde/Plzd_active_ligand/2'>Binding sites + ligand</scene> | ||
| + | |||
| + | <scene name='2rde/Pilz_domain/1'>Active sites</scene> (R136, R140, D162, S164, G167) | ||
</StructureSection> | </StructureSection> | ||
Current revision
VCA0042/plzD complexed with c-di-GMP
Introduction
The PDB code 2RDE represents a PilZ protein complexed with cyclic diguanylate monophosphate (c-di-GMP) in Vibrio cholera there is a molecular interaction involved in the regulation of bacterial biofilm formation and motility. The complex is formed by the binding of the small signaling molecule c-di-GMP to a protein domain known as PilZ. This interaction plays a crucial role in bacterial physiology and is implicated in various cellular processes. [1]
| |||||||||||
References
- ↑ Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007 Dec 12;26(24):5153-66. doi: 10.1038/sj.emboj.7601918. Epub 2007 Nov 22. PMID: 18034161; PMCID: PMC2140105.
- ↑ Pecina, Anna, et al. The Stand-Alone PilZ-Domain Protein MotL Specifically Regulates the Activity of the Secondary Lateral Flagellar System in Shewanella putrefaciens. Frontiers in Microbiology, vol. 12, 2021. Frontiers, https://www.frontiersin.org/articles/10.3389/fmicb.2021.668892.
- ↑ Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007 Dec 12;26(24):5153-66. doi: 10.1038/sj.emboj.7601918. Epub 2007 Nov 22. PMID: 18034161; PMCID: PMC2140105.
- ↑ Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007 Dec 12;26(24):5153-66. doi: 10.1038/sj.emboj.7601918. Epub 2007 Nov 22. PMID: 18034161; PMCID: PMC2140105.
- ↑ Hengge, R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7, 263–273 (2009). https://doi.org/10.1038/nrmicro2109
- ↑ Purificação, Aline Dias da, et al. The World of Cyclic Dinucleotides in Bacterial Behavior. Molecules, vol. 25, n. 10, Janeiro de 2020, p. 2462. www.mdpi.com, https://doi.org/10.3390/molecules25102462.
- ↑ Purificação, Aline Dias da, et al. The World of Cyclic Dinucleotides in Bacterial Behavior. Molecules, vol. 25, n. 10, Janeiro de 2020, p. 2462. www.mdpi.com, https://doi.org/10.3390/molecules25102462.
- ↑ Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007 Dec 12;26(24):5153-66. doi: 10.1038/sj.emboj.7601918. Epub 2007 Nov 22. PMID: 18034161; PMCID: PMC2140105.