User:Anjali Rabindran/Sandbox 1
From Proteopedia
< User:Anjali Rabindran(Difference between revisions)
| Line 23: | Line 23: | ||
=Mutations= | =Mutations= | ||
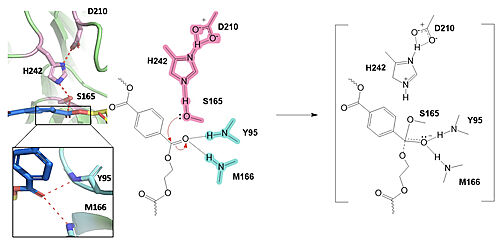
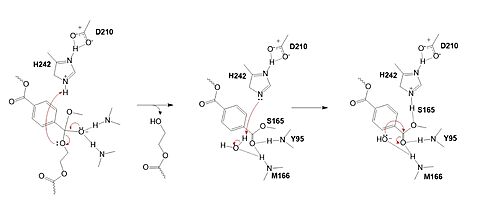
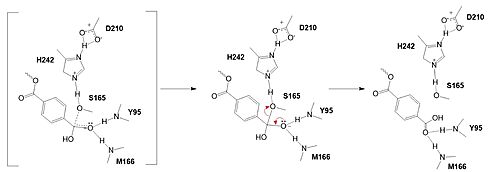
| - | Researchers have been investigating various mutations of PET hydrolase to enhance its catalytic ability. One group of researchers, Tournier et. al., have made mutations in the PET hydrolase active site. They identified the key residues involved in the catalytic mechanism by using a model of the <scene name='10/1075190/Ligand/ | + | Researchers have been investigating various mutations of PET hydrolase to enhance its catalytic ability. One group of researchers, Tournier et. al., have made mutations in the PET hydrolase active site. They identified the key residues involved in the catalytic mechanism by using a model of the <scene name='10/1075190/Ligand/4'>PET substrate</scene> (2-HE(MHET)₃) onto the enzyme (PDB ID 4EB0). The site, mainly a hydrophobic pocket, contained 11 residues targeted for mutagenesis. From this, they identified that the majority of enzymes' specific activity went down; however, the mutation of the <scene name='10/1075193/F243_wt/6'>wild type F243</scene> to either isoleucine or tryptophan increased specific activity. Four target mutations introduced into the PET Hydrolase by Tournier et. al. demonstrated improved catalytic efficiency and thermal stability compared to its wild-type structure. |
==F243W Mutation== | ==F243W Mutation== | ||
| - | The mutation of the F243 position to a tryptophan (<scene name='10/1075193/F243w_mutant/ | + | The mutation of the F243 position to a tryptophan (<scene name='10/1075193/F243w_mutant/7'>F243W</scene>) was selected for further analysis based on its enhanced catalytic activity in the depolymerization of Pf-PET. The W243 mutation was shown to improve the substrate's binding affinity and increase the enzyme’s activity compared to the wild-type enzyme. This was one of the few variants that exhibited higher activity, and it was further analyzed through differential scanning fluorimetry (DSF) to assess its stability. <ref name="Tournier et. al. 2020">PMID:32269349</ref> |
==F243I Mutation== | ==F243I Mutation== | ||
Similar to the W243 mutation, the <scene name='10/1075193/F243i/6'>F243I mutation</scene> was identified as a variant with improved depolymerization activity of Pf-PET. The I243 mutation in LCC led to better substrate interaction than the wild-type enzyme. After generating all possible variants, this mutation was among the few that exhibited 75% or more of the wild-type specific activity. As with the W243 mutation, DSF was used to determine the melting temperature and thermal stability, supporting the increased activity observed with this mutation. <ref name="Tournier et. al. 2020">PMID:32269349</ref> | Similar to the W243 mutation, the <scene name='10/1075193/F243i/6'>F243I mutation</scene> was identified as a variant with improved depolymerization activity of Pf-PET. The I243 mutation in LCC led to better substrate interaction than the wild-type enzyme. After generating all possible variants, this mutation was among the few that exhibited 75% or more of the wild-type specific activity. As with the W243 mutation, DSF was used to determine the melting temperature and thermal stability, supporting the increased activity observed with this mutation. <ref name="Tournier et. al. 2020">PMID:32269349</ref> | ||
Current revision
| |||||||||||