User:Letícia Oliveira Rojas Cruz/Sandbox 1
From Proteopedia
< User:Letícia Oliveira Rojas Cruz(Difference between revisions)
| (37 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | == | + | ==Angiotensin-converting enzyme 2 (ACE2) - (PDB 1R42)== |
| - | <StructureSection load='6m17' size=' | + | <StructureSection load='6m17' size='425' side='right' caption='PDB 6m17' scene='10/1083732/Ace2_completa/3'> |
| - | <scene name='10/1083732/Ace2_completa/ | + | <scene name='10/1083732/Ace2_completa/3'>Angiotensin-converting enzyme 2 (ACE2)</scene> is an important protein expressed in several human tissues, such as the heart and kidneys. It plays a key role in the Renin-Angiotensin-Aldosterone System - "RAAS" - by catalyzing the conversion of angiotensin II (Ang II), promoting blood pressure regulation. |
| - | Another function of ACE2 is related to SARS-CoV-2 infection, the virus responsible for the COVID-19 pandemic. In this context, ACE2 serves as the entry receptor for the virus <ref> | + | Another function of ACE2 is related to SARS-CoV-2 infection, the virus responsible for the COVID-19 pandemic. In this context, ACE2 serves as the entry receptor for the virus. <ref>PMID:33389262</ref> |
| - | + | [[Image:Protein ACE2 PDB 1r42.png|225px|right|thumb|Protein ACE2, [[1r42]]]] | |
| - | + | ||
| - | The | + | == General structure information == |
| + | The ACE2 protein gene has 40 kb and 18 exons. Its protein has 805 amino acids with a molecular weight of approximately 120 kDa. Here, we have a view of the <scene name='10/1083732/Ace2_completa/3'>full ACE2 protein</scene>, going from the '''{{Font color|blue|N-terminal}}''' to '''{{Font color|red|C-terminal}}''' region. Some other ways to represent the ACE2 protein include showing its <scene name='10/1083732/Ace2_alpha_e_beta/1'>secondary structure elements</scene>, such as '''{{Font color|deeppink|alpha helices}}''' and '''{{Font color|gold|beta sheets}}''', and mapping the '''polarity''' of its residues, differentiating <scene name='10/1083732/Ace2_completa_hydroph/1'>hydrophobic</scene> and <scene name='10/1083732/Ace2_completa_polar/1'>polar</scene> areas across its surface. | ||
| + | Besides its general aspects, the ACE2 protein can be divided into 4 portions: Peptidase Domain (Residues 19–615), Collectrin-like Domain (Residues 616–740), Transmembrane Domain (Residues 741–761) and Intracellular C-terminal Tail (Residues 762–805). | ||
| + | The '''<scene name='10/1083732/Peptidase_domain/2'>Peptidase Domain</scene>''' is responsible for the enzymatic activity of ACE2. This domain can be divided in two subdomains: <scene name='10/1083732/Subdominio_1_nterm/2'>Subdomain I</scene> (residues 19–400) and <scene name='10/1083732/Subdominio_2_cterm/2'>Subdomain II</scene> (residues 401–615). Together, they form a substrate-binding cleft, where is located the '''<scene name='10/1083732/Sitio_zinco_com_residuos/1'>catalytic site</scene>''', denominated '''HEXXH+E zinc-binding motif'''. Within this site, a '''{{Font color|lime|zinc-ion}}''' is associated with the residues '''His374''', '''His378''', and '''Glu402''', which are going to perform a nucleophilic attack on the peptide bond of the substrate, leading to its cleavage. '''<scene name='10/1083732/Peptidase_domain_hydrofobic/2'>Hydrophobic portions</scene>''' within each subdomain, composed mainly of nonpolar residues provide tertiary structural stability, maintaining the correct spatial arrangement of catalytic residues. In the animation, we can observe a concentration of the '''{{Font color|gray|hydrophobic residues}}''' towards the center of the molecule, while the '''{{Font color|orchid|polar}}''' ones are towards the outside part of the molecule. | ||
| - | = | + | An important region following the Peptidase Domain is the '''<scene name='10/1083732/Collectrin-like_domain/3'>Collectrin-like Domain</scene>''' (CLD), also described structurally as a ferredoxin-like fold or neck domain, comprising residues approximately between 616 and 726. This domain connects the Peptidase Domain to the Transmembrane Domain and is composed of '''{{Font color|gold|4 beta sheets}}''' and '''{{Font color|deeppink|4 alpha-helices}}''', arranged around a hydrophobic core. Its fold, characterized by a central beta-sheet flanked by alpha helices, plays a crucial role in stabilizing ACE2 dimers, which will be detailed later on this page. |
| - | == | + | The <scene name='10/1083732/Transmembrane_domain/1'>Transmembrane Domain</scene> is composed of a '''{{Font color|red|single alpha helix}}''', made of a majority of <scene name='10/1083732/Transmembrane_domain_hydrop/1'>hydrophobic residues</scene> (in '''{{Font color|gray|gray}}'''). This characteristic allows ACE2 to be anchored in the plasma membrane, due to its hydrophobic nature. |
| - | + | The final domain is the Intracellular C-terminal Tail but its function remains uncertain. | |
| - | == | + | == Physiological Function == |
| + | <scene name='10/1083732/Ace2_completa/3'>ACE2</scene> is a transmembrane glycoprotein that has an extracellular catalytic domain. It is classified as a hydrolase, more specifically a carboxypeptidase-type peptidase, responsible for the cleavage of peptide bonds at the C-terminal site. <ref>PMID:17897633</ref> | ||
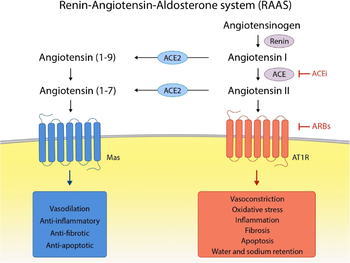
| + | The ACE2 acts to cleave a single C-terminal residue of some substrates. One of those substrates is Angiotensin II (Ang II), which is degrated into Angiotensin-(1-7); to a lesser extent, Angiotensin I (Ang I) is also a substrate, turning in into Angiotensin-(1-9). | ||
| + | The Renin-Angiotensin-Aldosterone System - "RAAS" -, in which ACE2 acts, is an important signaling pathway responsible for vascular homeostasis. | ||
| + | In general, Ang II, generated by the action of ACE on Ang I, and this one by the action of Renin on Angiotensinogen, acts on AT1 and AT2 receptors. Its main action is vasoconstriction (by AT1 receptor), increasing blood pressure. On the other hand, Angiotensin (1-7), generated from Ang II by ACE2, has the opposite function, leading to a decline in blood pressure through vasodilation and release of Nitric Oxide (NO) through its action on the MAS oncogene receptor, antagonizing the actions of Ang II. <ref>PMID:19461648</ref> | ||
| + | [[Image:RAAS.png|350px]] <ref>PMID:33389262</ref> | ||
| + | |||
| + | == Pathological Relevance == | ||
| + | The ACE2 protein became better known and widely publicized in 2020 due to its role in the infection by '''SARS-CoV-2''', the virus responsible for the Covid-19 pandemic. | ||
| + | In this disease, ACE2 acts as a viral receptor, to which the Spike (S) viral protein binds, promoting the entry of the virus into the host cell. The specific region of the S protein responsible for binding to the ACE2 receptor is called '''receptor-binding domain (RBD)''', and is located in the S1 subunit of the Spike protein. <ref>PMID:32142651</ref> | ||
| + | |||
| + | == Structure highlights<ref>PMID:32132184</ref>== | ||
| + | === ACE2-B0AT1 Complex === | ||
| + | '''''Association with B0AT1''''' | ||
| + | |||
| + | The ACE2 protein can associate with the neutral amino acid transporter <scene name='10/1078774/B0at1_zoom2/1'>B0AT1</scene>, also known as SLC6A19. In this context, ACE2 first forms a <scene name='10/1083732/Dimer_ace2_final/2'>homodimer</scene>, where two ACE2 molecules interact side by side. Each ACE2 monomer then binds to one B0AT1 molecule. This results in a complex composed of '''{{Font color|darkviolet|two ACE2}}''' and '''{{Font color|salmon|two B0AT1}}''' molecules, which is commonly described as a <scene name='10/1083732/Ace2_and_b0at1/2'>dimer of heterodimers</scene>. This association is essential for the transport of neutral amino acids in intestinal cells and the anchoring of ACE2 in the cell membrane, keeping the catalytic site facing the extracellular environment. Although ACE2 dimerization occurs independently of B0AT1, the transporter plays a stabilizing role by interacting with the Collectrin-like Domain. | ||
| + | |||
| + | This ACE2-B0AT1 complex is anchored to the cell plasma membrane, keeping the ACE2 protein in an extracellular environment. The intermembrane region has apolar characteristics, since it must interact with the hydrophobic tails of the phospholipids. Analyzing the <scene name='10/1083732/Polaridade_2/2'>polarity</scene> of the complex, an '''{{Font color|gray|apolar region}}''' is observed in the B0AT1 region, showing that it is in this region where the complex is in contact with the plasma membrane. The '''{{Font color|orchid|polar regions}}''' (CLD and PD domains of ACE2) are located in the external region, without contact with the apolar region of the phospholipids. | ||
| + | |||
| + | === ACE2 Dimerization === | ||
| + | ACE2 protein dimerization occurs independently of B0AT1 and involves both the Collectrin-like Domain (CLD) and regions from the Peptidase Domain (PD). | ||
| + | |||
| + | '''''CLD Domain Interaction''''' | ||
| + | |||
| + | In this region, there is an extensive network of polar interactions that stabilizes the ACE2 dimer. The most actively involved amino acid residues are between 636 and 658 and between 708 and 717, corresponding to the second and fourth helices of the CLD domain, respectively. | ||
| + | |||
| + | <scene name='10/1078774/Interacao_cation/2'>Polar cation-pi interactions</scene> occur between the amino acids '''{{Font color|slateblue|Arg652}}''' and '''{{Font color|turquoise|Tyr641}}''' of the opposite ACE2 molecule, and between '''{{Font color|slateblue|Arg710}}''' and '''{{Font color|turquoise|Tyr633}}'''. Additionally, <scene name='10/1078774/Lig_hidrogenio/1'>hydrogen bonds</scene> help stabilize the dimer interface: '''{{Font color|slateblue|Arg652}}''' forms a hydrogen bond with '''{{Font color|slateblue|Arg638}}''', which further interacts with '''{{Font color|slateblue|Gln653}}''', linked to '''{{Font color|turquoise|Asn636}}'''. Also, '''{{Font color|slateblue|Arg710}}''' forms a hydrogen bond with '''{{Font color|turquoise|Asn639}}''', and '''{{Font color|turquoise|Arg716}}''' connects with '''{{Font color|slateblue|Ser709}}''' and '''{{Font color|slateblue|Asp713}}'''. | ||
| + | |||
| + | '''''Domain PD Interation''''' | ||
| + | In addition to the CLD, the Peptidase Domain (PD) also contributes to ACE2 dimerization. The amino acids '''{{Font color|slateblue|Cys133}}''', '''{{Font color|slateblue|Asn134}}''', '''{{Font color|slateblue|Asp136}}''', '''{{Font color|slateblue|Asn137}}''', '''{{Font color|slateblue|Gln139}}''', '''{{Font color|slateblue|Glu140}}''', and '''{{Font color|slateblue|Cys141}}''' form a <scene name='10/1083732/Loop_pd/2'>loop</scene> region within the PD. The two cysteines ('''{{Font color|slateblue|Cys133}}''' and '''{{Font color|slateblue|Cys141}}''') establish a disulfide bond, which stabilizes the loop conformation, supported by additional intraloop polar interactions. Furthermore, '''{{Font color|slateblue|Gln139}}''' forms a polar interaction with '''{{Font color|turquoise|Gln175}}''' from the other ACE2 monomer. | ||
| + | |||
| + | === SARS-CoV-2 Binding === | ||
| + | '''{{Font color|crimson|ACE2}}''' <scene name='10/1083732/Sars_cov_2/2'>binds</scene> to the '''{{Font color|darkmagenta|Spike (S)}}''' glycoprotein of the SARS-CoV-2 virus, promoting its internalization into the cell. More specifically, binding occurs between the PD subunit of ACE2 and the receptor binding domain (RBD) of the S1 subunit of the S protein. Each PD binds to an RBD by polar bonds, with one ACE2 dimer accommodating two S protein trimers. | ||
</StructureSection> | </StructureSection> | ||
| + | |||
== References == | == References == | ||
<references/> | <references/> | ||
| + | |||
| + | |||
| + | OUDIT, G. Y. et al. Angiotensin-converting enzyme 2—at the heart of the COVID-19 pandemic. Cell, v. 186, n. 5, p. 906–922, mar. 2023. | ||
Current revision
Angiotensin-converting enzyme 2 (ACE2) - (PDB 1R42)
| |||||||||||
References
- ↑ Beyerstedt S, Casaro EB, Rangel ÉB. COVID-19: angiotensin-converting enzyme 2 (ACE2) expression and tissue susceptibility to SARS-CoV-2 infection. Eur J Clin Microbiol Infect Dis. 2021 May;40(5):905-919. PMID:33389262 doi:10.1007/s10096-020-04138-6
- ↑ Lambert DW, Hooper NM, Turner AJ. Angiotensin-converting enzyme 2 and new insights into the renin-angiotensin system. Biochem Pharmacol. 2008 Feb 15;75(4):781-6. PMID:17897633 doi:10.1016/j.bcp.2007.08.012
- ↑ Iwai M, Horiuchi M. Devil and angel in the renin-angiotensin system: ACE-angiotensin II-AT1 receptor axis vs. ACE2-angiotensin-(1-7)-Mas receptor axis. Hypertens Res. 2009 Jul;32(7):533-6. PMID:19461648 doi:10.1038/hr.2009.74
- ↑ Beyerstedt S, Casaro EB, Rangel ÉB. COVID-19: angiotensin-converting enzyme 2 (ACE2) expression and tissue susceptibility to SARS-CoV-2 infection. Eur J Clin Microbiol Infect Dis. 2021 May;40(5):905-919. PMID:33389262 doi:10.1007/s10096-020-04138-6
- ↑ Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Muller MA, Drosten C, Pohlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020 Apr 16;181(2):271-280.e8. doi: 10.1016/j.cell.2020.02.052. Epub 2020, Mar 5. PMID:32142651 doi:http://dx.doi.org/10.1016/j.cell.2020.02.052
- ↑ Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science. 2020 Mar 4. pii: science.abb2762. doi: 10.1126/science.abb2762. PMID:32132184 doi:http://dx.doi.org/10.1126/science.abb2762
OUDIT, G. Y. et al. Angiotensin-converting enzyme 2—at the heart of the COVID-19 pandemic. Cell, v. 186, n. 5, p. 906–922, mar. 2023.