Journal:Acta Cryst D:S2059798323007672
From Proteopedia
(Difference between revisions)

| (3 intermediate revisions not shown.) | |||
| Line 10: | Line 10: | ||
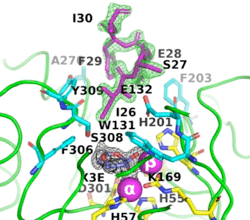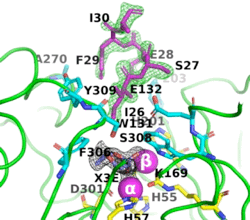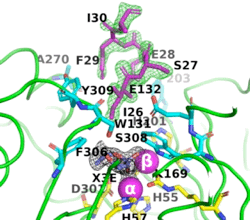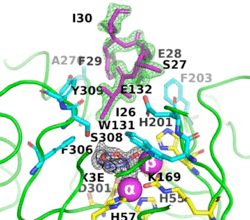
The image on the left shows the electron density (in green) of an omit map with the octapeptide omitted (contoured at 3 sigma). The cyclic compound, X3E, presumably carried over from the protein expression and purification process, is seen in the active site, and the electron density (in black) corresponds to a 2Fo-Fc map (contoured at 1 sigma). It is clear that the particular crystallization conditions and space group can have an enormous impact on whether a ligand binds to a protein. | The image on the left shows the electron density (in green) of an omit map with the octapeptide omitted (contoured at 3 sigma). The cyclic compound, X3E, presumably carried over from the protein expression and purification process, is seen in the active site, and the electron density (in black) corresponds to a 2Fo-Fc map (contoured at 1 sigma). It is clear that the particular crystallization conditions and space group can have an enormous impact on whether a ligand binds to a protein. | ||
| - | <scene name='99/992467/C23m_1/ | + | The <scene name='99/992467/C23m_1/3'>active-site region of C23M_1</scene> (PDB-ID [[8p7u]]), shows the active-site residues in yellow. the two Zn<sup>+2</sup> ions as magenta spheres. |
| - | <scene name='99/992467/C23_2/ | + | <scene name='99/992467/C23_2/10'>O-isopropyl methylphosphonic acid was observed in C23_2</scene> (PDB-ID [[8p7u]]). The six residues that bind to the two Zn<sup>2+</sup> ions are shown as ball-and-stick figures, with carbons coloured yellow, nitrogens blue, oxygens red, and phosphorus atoms orange. The interatomic distances observed between the oxygens of the P-O in all OP acid products are 1.9-2.0 Å. |
Overall, this study provides valuable insights into the challenges and considerations involved in studying the crystal structures of ligand-protein complexes, highlighting the importance of careful experimental design and rigorous data analysis in ensuring the accuracy and reliability of the resulting PTE-OP structures obtained. | Overall, this study provides valuable insights into the challenges and considerations involved in studying the crystal structures of ligand-protein complexes, highlighting the importance of careful experimental design and rigorous data analysis in ensuring the accuracy and reliability of the resulting PTE-OP structures obtained. | ||
Current revision
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.