|
|
| (121 intermediate revisions not shown.) |
| Line 1: |
Line 1: |
| - | {{STRUCTURE_2nmz| PDB=2nmz | SCENE= HIV-1_protease/2nmz/3 }}
| + | <StructureSection load='2nmz' size='350' side='right' background='none' scene='User:David_Canner/Sandbox_HIV/Opening/2' caption='Structure of HIV-1 Protease (PDB code [[2nmz]])'> |
| - | HIV is a notoriously lethal virus that is known to cause AIDS. There currently is no cure or vaccine. But, scientists have discovered treatments that can slow progression of the HIV virus, thanks in large part to our understanding of the structure of [[HIV-1 protease]], seen here on the right in complex with a potent drug used for slowing the progression of HIV, <scene name='HIV-1_protease/2nmz_saquinavir/1'>Saquinavir</scene> (PDB entry [[2nmz]]).
| + | __TOC__ |
| | | | |
| - | HIV-1 protease is a protein made by the HIV virus that is crucial to the virus's infectious capacity. The virus makes certain proteins that need to be cleaved, or cut, in order to transform into mature, fully-functional proteins that can allow the virus to infect new cells. HIV-1 protease is responsible for cleaving these nascent proteins into their mature form. | |
| | | | |
| - | Looking at the structure of HIV-1 protease, we see that the protein is composed of <scene name='HIV-1_protease/2nmz_symmetric/2'>two symmetrically related subunits</scene>, shown here in [[cartoon backbone representation]] to highlight [[secondary structure]]. Each subunit consists of the same small chain of only 99 amino acids. The subunits come together in such as way as to <scene name='HIV-1_protease/2nmz_tunnel/1'>form a tunnel where they meet</scene>, shown here in [[spacefilling representation]] to showcase the physical surface of the protein. The protein to-be-cleaved sits in this tunnel. In the middle of the tunnel is the <scene name='HIV-1_protease/2nmz_triads/1'>active site</scene> of the protease: <scene name='HIV-1_protease/2nmz_triadslabeled/2'>two Asp-Thr-Gly catalytic triads</scene> (residue numbers 25, 26, and 27 on one chain and 125, 126, and 127 on the second). <scene name='HIV-1_protease/2nmz_aspslabeled/1'>The two Asp's</scene> act as the main catalytic residues in the active site and use a water molecule to help break the protein chain that binds in the tunnel.
| + | ==Function== |
| | + | [[Human Immunodeficiency Virus]] (HIV) is the cause of Acquired Immunodeficiency Syndrome (AIDS). HIV directs the synthesis of several polyproteins, which each consist of several tandemly linked proteins. The maturation of the virus to its infectious form requires that these polyproteins be cleaved to their component proteins. <scene name='User:David_Canner/Sandbox_HIV/Opening/2'>HIV-1 protease</scene>, a homodimeric enzyme, is responsible for doing so and is therefore crucial to the virus's infectious capacity.<br /> |
| | + | HIV exists in two types '''HIV-1''' and '''HIV-2'''. HIV-2 infects ca. 30% of AIDS patients vs. 70% infected by HIV-1<ref>PMID:22238126</ref>.<br /> |
| | + | '''FIV''' is Feline Immunodeficiency virus protease.<br /> |
| | + | '''SIV''' is Simian Immunodeficiency virus protease.<br /> |
| | | | |
| - | Saquinavir was the the first FDA protease inhibitor approved for the treatment of HIV. It inhibits HIV-1 protease by <scene name='HIV-1_protease/2nmz_saquinavir_spacefill/1'>binding tightly to the active site tunnel</scene>, thus preventing the protease from cleaving any protein chains. You may be wondering how a protein-to-be cleaved makes its way into the active-site tunnel to begin with -- afterall, the tunnel does not seem so accessible. The key are the two flexible flaps on the top of the tunnel that can <scene name='HIV-1_protease/Hiv1_protease_morph/3'>move</scene> (large scene, takes a while to load) to allow proteins to enter the tunnel. A <scene name='HIV-1_protease/Hiv1_p_morph_sp/1'>spacefill view of the flexible flaps</scene> is also illuminating, as the change in the accessibility of the tunnel becomes more obvious. This movement of the flexible flaps is simulated by morphing between two crystal structures, the first being the native HIV-1 protease structure with no inhibitor bound (PDB entry [[1hhp]]) and the second being the HIV-1 protease complexed with Saquinavir.
| + | See also:[[Flaps Morph for HIV Protease]]. |
| | | | |
| - | Other drugs used to treat patients infected with the HIV virus include (PDB entry [[1hsg]]), Ritonavir (PDB entry [[1hxw]]), and Nelfinavir (PDB entry [[1ohr]]).
| + | ==Structure of HIV-1 Protease== |
| | + | The X-ray structure of HIV-1 protease<ref>PMID:2548279</ref><ref>PMID:2682266</ref> reveals that it is composed of <scene name='User:David_Canner/Sandbox_HIV/Identical_subunits/1'>two symmetrically related subunits</scene>, each consisting of 99 amino acid residues. The subunits come together in such as way as to <scene name='User:David_Canner/Sandbox_HIV/Tunnel/1'>form a tunnel where they meet</scene>. This tunnel is of critical importance because the active site of the protease is located in its interior. The active site consists of <scene name='User:David_Canner/Sandbox_HIV/Catalytic_triad/3'> two Asp-Thr-Gly conserved sequences</scene>, making it a member of the aspartyl protease family. The two Asp's are <scene name='User:David_Canner/Sandbox_HIV/Catalytic_asp/1'>essential catalytic residues</scene> either interact with the incoming water OR protonate the carbonyl to make the carbon more electrophilic for the incoming <scene name='31/315240/Saquinavir_cat_water/2'>water</scene>. You may be wondering how a polyprotein makes its way into the active-site tunnel, as the<scene name='User:David_Canner/Sandbox_HIV/Narrow_tunnel/1'> tunnel appears to be too narrow </scene> to admit it. The key is the two flexible flaps on the top of the tunnel that <scene name='User:David_Canner/Sandbox_HIV/Hiv_tunnel_morph/3'>move to allow proteins </scene>to enter the tunnel. The flaps <scene name='User:David_Canner/Sandbox_HIV/Hiv_tunnel_morph_flaps/2'>undergo a dramatic movement</scene>, shifting from an open to a closed conformation to bind the target in an appropriate conformation for cleavage. This is more clearly seen at [[Flaps Morph for HIV Protease]]. |
| | | | |
| - | {{Clear}}
| + | ==Medical Implications== |
| | + | There currently is no cure or vaccine against HIV. Researchers, however, have discovered treatments that can halt and even reverse the progression of AIDS, due in large part to our understanding of the structure of HIV-1 protease. <scene name='User:David_Canner/Sandbox_HIV/Saquinavir/4'>Saquinavir</scene> ([[Invirase]]) was the first protease inhibitor approved by the FDA for the treatment of HIV. It inhibits HIV protease by <scene name='User:David_Canner/Sandbox_HIV/Saquinavir_tunnel/1'>binding tightly in the active site tunnel</scene>, preventing the binding of polyproteins. Its chemical structure mimics the tetrahedral intermediate of the hydrolytic reaction, thereby <scene name='User:David_Canner/Sandbox_HIV/Saquinavir_cat/3'>interacting strongly with the catalytic Asp residues</scene>.<ref>PMID:17243183</ref> Saquinavir is essentially an uncleavable ligand, as indicated by the <scene name='User:David_Canner/Sandbox_HIV/Hiv_morph2/9'> similar conformational changes in the protease flaps </scene> on binding saquinavir or a polypeptide. Resistance to saquinavir is due to alterations in the HIV protease sequence, including the mutation of <scene name='31/315240/Saquinavir_mut/1'>Leu 10 and Ile 50</scene><ref>PMID: 8969180</ref>. Drugs used to treat HIV infection that inhibit <scene name='User:David_Canner/Sandbox_HIV/Inhibitor_intro/1'>HIV protease</scene> include <scene name='User:David_Canner/Sandbox_HIV/Indinavir/2'>Indinavir </scene> ([[Crixivan]]), <scene name='User:David_Canner/Sandbox_HIV/Ritonavir/1'>Ritonavir</scene> ([[Norvir]]), [[Saquinavir]], [[Tipranavir]], [[Amprenavir]] (Agenerase), [[Atazanavir]] (Rayataz), [[Darunavir]] (Prezista), [[Fosamprenavir]] (Lexiva or Telzir), [[Lopinavir]] (Kaletra), [[Nelfinavir]] (Viracept) and <scene name='User:David_Canner/Sandbox_HIV/Nelfinavir/2'>Nelfinavir</scene> ([[Viracept]]). |
| | | | |
| - | ==References==
| + | See also [[Treatments:HIV Protease Inhibitor Pharmacokinetics References]] |
| - | *Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir., Tie Y, Kovalevsky AY, Boross P, Wang YF, Ghosh AK, Tozser J, Harrison RW, Weber IT, Proteins. 2007 Apr 1;67(1):232-42. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/17243183 17243183]
| + | |
| - | *The three-dimensional structure of the aspartyl protease from the HIV-1 isolate BRU., Spinelli S, Liu QZ, Alzari PM, Hirel PH, Poljak RJ, Biochimie. 1991 Nov;73(11):1391-6. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/1799632 1799632]
| + | |
| | | | |
| - | ==Links== | + | == Structural Insights into the South African HIV-1 Subtype C Protease: Impact of hinge region dynamics and flap flexibility in drug resistance <ref>doi 10.1080/07391102.2012.736774</ref>== |
| - | * HIV-1 Protease featured in [[User:David S. Goodsell | David S. Goodsell's]] [http://mgl.scripps.edu/people/goodsell/pdb/pdb6/pdb6_1.html Molecule of the Month]
| + | The current study reports on the apo crystal structure of the <scene name='Journal:JBSD:36/Cv/3'>South African HIV-1 subtype C protease (C-SA PR)</scene>. Structure of <scene name='Journal:JBSD:36/Cv/4'>unbound HIV-1 PR</scene> with the active site triplet (D25, T26 and G27) shown in ball-and-stick representation, <font color='magenta'><b>hinge region in magenta (residues 35–42 and 57–61)</b></font>, and <span style="color:cyan;background-color:black;font-weight:bold;">flap region (residues 46–54) in cyan</span>. The relevance of this study cannot be underestimated because South Africa is at the epicenter of the HIV/AIDS pandemic. A detailed understanding of the molecular interactions between the drug and its target is required if we are to improve the design of protease inhibitors (PIs). Our study indicated that the loss of a salt bridge between <scene name='Journal:JBSD:36/Cv/5'>residues E35 and R57</scene> at the hinge region affects the flap dynamics of the apo C-SA PR which may reduce the affinity and, therefore, the efficacy of the current protease inhibitors toward the C-SA PR (<span style="color:deeppink;background-color:black;font-weight:bold;">subtype C-SA PR is in deeppink</span>, [[3u71]] and <span style="color:yellow;background-color:black;font-weight:bold;">subtype B PR is in yellow</span>, [[2pc0]]). <scene name='Journal:JBSD:36/Cv/6'>Structural alignment</scene> of of the <span style="color:deeppink;background-color:black;font-weight:bold;">C-SA PR (deep pink</span>, PDB ID: [[3u71]]), <span style="color:yellow;background-color:black;font-weight:bold;">consensus subtype B PR (yellow</span>, PDB ID: [[2pc0]]), and <span style="color:wheat;background-color:black;font-weight:bold;">subtype B-MDR PR (color wheat</span>, PDB ID: [[1rp1]]) reveals that the PRs under investigation do not differ significantly. The crystal structure of the C-SA PR will serve as a foundation to improve the rational design of PIs which will have a greater impact on anti-retroviral chemotherapy in sub-Saharan Africa. |
| - | * HIV-1 Protease in [http://en.wikipedia.org/wiki/HIV-1_protease Wikipedia]
| + | |
| | | | |
| | + | ==3D Printed Physical Model of HIV Protease== |
| | | | |
| - | ''This page is not a fully developed page, but was created as an example for the press release of the [http://genomebiology.com/2008/9/8/R121 Proteopedia article] in the open-access journal Genome Biology. Please expand this page with additional information and references.''
| + | Shown below are 3D printed physical models of HIV Protease. Both versions are shown in alpha carbon format, with select side chains shown colored by element, with carbon gray, nitrogen blue, oxygen red and sulfur yellow. Both models have been designed with precisely embedded magnets that allow the two chains to pull apart into individual pieces. |
| | + | |
| | + | [[Image:hivProtease1_centerForBioMolecularModeling_crop.jpg|230px]] [[Image:800px-HivProtease2 centerForBioMolecularModeling Crop.jpg|230px]] |
| | + | |
| | + | The MSOE Center for BioMolecular Modeling |
| | + | |
| | + | [[Image:CbmUniversityLogo.jpg | left | 150px]] |
| | + | |
| | + | The [http://cbm.msoe.edu MSOE Center for BioMolecular Modeling] uses 3D printing technology to create physical models of protein and molecular structures, making the invisible molecular world more tangible and comprehensible. To view more protein structure models, visit our [http://cbm.msoe.edu/educationalmedia/modelgallery/ Model Gallery]. |
| | + | |
| | + | |
| | + | ==HIV Protease Movie== |
| | + | <br> |
| | + | <br> HIV Protease Movie by Warren L. DeLano (made via PyMol) |
| | + | <br> |
| | + | <html5media height=“500” width=“1000”>https://www.youtube.com/embed/iSeVYYDvLCk</html5media> |
| | + | |
| | + | ==Additional resources== |
| | + | *''Aids Before Protease Inhibitors'' and ''HIV Protease Inhibitors: A Breakthrough'' at [[Molecular Playground/HIV Protease Inhibitor|HIV Protease Inhibitor]]. |
| | + | *[[Flaps Morph for HIV Protease]] |
| | + | *[[HIV Protease Inhibitor Pharmacokinetics]] |
| | + | *[[Treatments:HIV Protease Inhibitor Pharmacokinetics References]] |
| | + | *[[HIV Protease Inhibitor Resistance Profile]] |
| | + | *[[HIV Protease Resistance]] |
| | + | *[[Viability of a drug-resistant HIV-1 protease mutant]] |
| | + | *[[HIV and accessory proteins]] |
| | + | *[[Treatments:HIV Protease Inhibitor Pharmacokinetics References]] |
| | + | *[[Group:SMART:HIV-1 Subtype C Protease]] |
| | + | *[[Human Immunodeficiency Virus]] |
| | + | *[[Ann Taylor/HIV Protease]] |
| | + | *[[Virus protease]] |
| | + | *[[HIV-1 protease]] |
| | + | *[[Protease]] |
| | + | *[[Viability of a drug-resistant HIV-1 protease mutant]] |
| | + | * Structural Insights into the South African HIV-1 Subtype C Protease: Impact of hinge region dynamics and flap flexibility in drug resistance <ref>doi 10.1080/07391102.2012.736774</ref> |
| | + | * [http://cdn.rcsb.org/pdb101/learn/resources/structural-biology-of-hiv/index.html Structural Biology of HIV], an interactive Flash graphic of the virion with explanations of its components. |
| | + | |
| | + | ==Immunodeficiency virus protease 3D structures== |
| | + | [[Immunodeficiency virus protease 3D structures]] |
| | + | |
| | + | ==References== |
| | + | <references/> |
| | + | </StructureSection> |
| | + | [[Category:Topic Page]] |
| | + | [[Category:3D printer files]] |
|
Function
Human Immunodeficiency Virus (HIV) is the cause of Acquired Immunodeficiency Syndrome (AIDS). HIV directs the synthesis of several polyproteins, which each consist of several tandemly linked proteins. The maturation of the virus to its infectious form requires that these polyproteins be cleaved to their component proteins. , a homodimeric enzyme, is responsible for doing so and is therefore crucial to the virus's infectious capacity.
HIV exists in two types HIV-1 and HIV-2. HIV-2 infects ca. 30% of AIDS patients vs. 70% infected by HIV-1[1].
FIV is Feline Immunodeficiency virus protease.
SIV is Simian Immunodeficiency virus protease.
See also:Flaps Morph for HIV Protease.
Structure of HIV-1 Protease
The X-ray structure of HIV-1 protease[2][3] reveals that it is composed of , each consisting of 99 amino acid residues. The subunits come together in such as way as to . This tunnel is of critical importance because the active site of the protease is located in its interior. The active site consists of , making it a member of the aspartyl protease family. The two Asp's are either interact with the incoming water OR protonate the carbonyl to make the carbon more electrophilic for the incoming . You may be wondering how a polyprotein makes its way into the active-site tunnel, as the to admit it. The key is the two flexible flaps on the top of the tunnel that to enter the tunnel. The flaps , shifting from an open to a closed conformation to bind the target in an appropriate conformation for cleavage. This is more clearly seen at Flaps Morph for HIV Protease.
Medical Implications
There currently is no cure or vaccine against HIV. Researchers, however, have discovered treatments that can halt and even reverse the progression of AIDS, due in large part to our understanding of the structure of HIV-1 protease. (Invirase) was the first protease inhibitor approved by the FDA for the treatment of HIV. It inhibits HIV protease by , preventing the binding of polyproteins. Its chemical structure mimics the tetrahedral intermediate of the hydrolytic reaction, thereby .[4] Saquinavir is essentially an uncleavable ligand, as indicated by the on binding saquinavir or a polypeptide. Resistance to saquinavir is due to alterations in the HIV protease sequence, including the mutation of [5]. Drugs used to treat HIV infection that inhibit include (Crixivan), (Norvir), Saquinavir, Tipranavir, Amprenavir (Agenerase), Atazanavir (Rayataz), Darunavir (Prezista), Fosamprenavir (Lexiva or Telzir), Lopinavir (Kaletra), Nelfinavir (Viracept) and (Viracept).
See also Treatments:HIV Protease Inhibitor Pharmacokinetics References
Structural Insights into the South African HIV-1 Subtype C Protease: Impact of hinge region dynamics and flap flexibility in drug resistance [6]
The current study reports on the apo crystal structure of the . Structure of with the active site triplet (D25, T26 and G27) shown in ball-and-stick representation, hinge region in magenta (residues 35–42 and 57–61), and flap region (residues 46–54) in cyan. The relevance of this study cannot be underestimated because South Africa is at the epicenter of the HIV/AIDS pandemic. A detailed understanding of the molecular interactions between the drug and its target is required if we are to improve the design of protease inhibitors (PIs). Our study indicated that the loss of a salt bridge between at the hinge region affects the flap dynamics of the apo C-SA PR which may reduce the affinity and, therefore, the efficacy of the current protease inhibitors toward the C-SA PR (subtype C-SA PR is in deeppink, 3u71 and subtype B PR is in yellow, 2pc0). of of the C-SA PR (deep pink, PDB ID: 3u71), consensus subtype B PR (yellow, PDB ID: 2pc0), and subtype B-MDR PR (color wheat, PDB ID: 1rp1) reveals that the PRs under investigation do not differ significantly. The crystal structure of the C-SA PR will serve as a foundation to improve the rational design of PIs which will have a greater impact on anti-retroviral chemotherapy in sub-Saharan Africa.
3D Printed Physical Model of HIV Protease
Shown below are 3D printed physical models of HIV Protease. Both versions are shown in alpha carbon format, with select side chains shown colored by element, with carbon gray, nitrogen blue, oxygen red and sulfur yellow. Both models have been designed with precisely embedded magnets that allow the two chains to pull apart into individual pieces.
 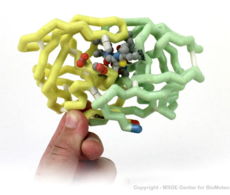
The MSOE Center for BioMolecular Modeling
The MSOE Center for BioMolecular Modeling uses 3D printing technology to create physical models of protein and molecular structures, making the invisible molecular world more tangible and comprehensible. To view more protein structure models, visit our Model Gallery.
HIV Protease Movie
HIV Protease Movie by Warren L. DeLano (made via PyMol)
Additional resources
Immunodeficiency virus protease 3D structures
Immunodeficiency virus protease 3D structures
References
- ↑ Tie Y, Wang YF, Boross PI, Chiu TY, Ghosh AK, Tozser J, Louis JM, Harrison RW, Weber IT. Critical differences in HIV-1 and HIV-2 protease specificity for clinical inhibitors. Protein Sci. 2012 Mar;21(3):339-50. doi: 10.1002/pro.2019. Epub 2012 Jan 24. PMID:22238126 doi:10.1002/pro.2019
- ↑ Wlodawer A, Miller M, Jaskolski M, Sathyanarayana BK, Baldwin E, Weber IT, Selk LM, Clawson L, Schneider J, Kent SB. Conserved folding in retroviral proteases: crystal structure of a synthetic HIV-1 protease. Science. 1989 Aug 11;245(4918):616-21. PMID:2548279
- ↑ Lapatto R, Blundell T, Hemmings A, Overington J, Wilderspin A, Wood S, Merson JR, Whittle PJ, Danley DE, Geoghegan KF, et al.. X-ray analysis of HIV-1 proteinase at 2.7 A resolution confirms structural homology among retroviral enzymes. Nature. 1989 Nov 16;342(6247):299-302. PMID:2682266 doi:http://dx.doi.org/10.1038/342299a0
- ↑ Tie Y, Kovalevsky AY, Boross P, Wang YF, Ghosh AK, Tozser J, Harrison RW, Weber IT. Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir. Proteins. 2007 Apr 1;67(1):232-42. PMID:17243183 doi:10.1002/prot.21304
- ↑ Maschera B, Darby G, Palu G, Wright LL, Tisdale M, Myers R, Blair ED, Furfine ES. Human immunodeficiency virus. Mutations in the viral protease that confer resistance to saquinavir increase the dissociation rate constant of the protease-saquinavir complex. J Biol Chem. 1996 Dec 27;271(52):33231-5. PMID:8969180
- ↑ Naicker P, Achilonu I, Fanucchi S, Fernandes M, Ibrahim MA, Dirr HW, Soliman ME, Sayed Y. Structural insights into the South African HIV-1 subtype C protease: impact of hinge region dynamics and flap flexibility in drug resistance. J Biomol Struct Dyn. 2012 Nov 12. PMID:23140382 doi:10.1080/07391102.2012.736774
- ↑ Naicker P, Achilonu I, Fanucchi S, Fernandes M, Ibrahim MA, Dirr HW, Soliman ME, Sayed Y. Structural insights into the South African HIV-1 subtype C protease: impact of hinge region dynamics and flap flexibility in drug resistance. J Biomol Struct Dyn. 2012 Nov 12. PMID:23140382 doi:10.1080/07391102.2012.736774
|



