Acetylcholinesterase
From Proteopedia
| Line 35: | Line 35: | ||
{{Clear}} | {{Clear}} | ||
| + | <applet load='ACHE2.pdb' size='500' frame='true' align='left' | ||
| + | scene='AChE_inhibitors_and_substrates/Com_view_tacrine/1' /> | ||
| + | [http://en.wikipedia.org/wiki/Tacrine Tacrine]. In the X-ray crystal structure of ''Tc''AChE/<scene name='AChE_inhibitors_and_substrates/Com_view_tacrine/2'>tacrine</scene> complex which was determined at 2.8 Å resolution, the tacrine is seen <font color='magenta'><b>(magenta)</b></font> bound in the active site of ''Tc''AChE ([[1acj]]) . <font color='gray'><b>ACh (gray)</b></font> is shown for comparison. | ||
| + | {{Clear}} | ||
| + | <applet load='1gqr' size='500' frame='true' align='right' | ||
| + | scene='1gqr/Com_view/1' /> | ||
| + | |||
| + | [http://en.wikipedia.org/wiki/Rivastigmine Rivastigmine (Exelon)] is a [http://en.wikipedia.org/wiki/Carbamate carbamate] inhibitor of AChE, and it is currenly used in therapy of [http://en.wikipedia.org/wiki/Alzheimer's_disease Alzheimer's disease]. Rivastigmine (colored yellow) interacts with ''Tc''AChE <font color='lime'><b>(colored lime)</b></font> at the <scene name='1gqr/Active_site/4'>active-site gorge</scene> ([[1gqr]]). The carbamyl moiety of rivastigmine is <scene name='1gqr/Active_site/9'>covalently bound</scene> to the active-site S200 Oγ. The second part of rivastigmine (the leaving group), NAP ((−)-S-3-[1-(dimethylamino)ethyl]phenol) is also held in the active-site gorge, but it is <scene name='1gqr/Active_site/6'>separated</scene> from the carbamyl moiety, hence, carbamylation took place. The <scene name='1gqr/Active_site/7'>crystal structure</scene> of ''Tc''AChE/<font color='magenta'><b>NAP (colored magenta)</b></font> is known ([[1gqs]]). The <font color='violet'><b>''Tc''AChE active-site residues</b></font> which are interacting with NAP are <font color='violet'><b>colored violet</b></font>. NAP is located in a similar region of ''Tc''AChE active site, but with different orientation than that of the NAP part (colored yellow) in the ''Tc''AChE/rivastigmine complex. Only H440 and F330 significantly change their side-chain conformations. <scene name='1gqr/Active_site/8'>Overlap</scene> of the ''Tc''AChE active sites in 4 different structures (<font color='lime'><b>''Tc''AChE</b></font>/rivastigmine ([[1gqr]]), <font color='violet'><b>''Tc''AChE</b></font>/<font color='magenta'><b>NAP</b></font> ([[1gqs]]), <font color='cyan'><b>native ''Tc''AChE</b></font> ([[2ace]]), and ''Tc''AChE/'''VX''' ([[1vxr]], ''Tc''AChE colored white and VX black) reveals that the conformation of H440 in the ''Tc''AChE/NAP structure is very similar its conformation in the native ''Tc''AChE ([[2ace]]), but the distance between H440 Nδ and E327 Oε is significantly longer in the ''Tc''AChE/rivastigmine and the ''Tc''AChE/'''VX''' complexes. This structural change disrupts the [http://en.wikipedia.org/wiki/Catalytic_triad catalytic triad] consisting of S200, E327, H440. This could explain the very slow kinetics of AChE reactivation after its inhibition by rivastigmine. | ||
{{Clear}} | {{Clear}} | ||
Revision as of 09:54, 1 November 2010
|
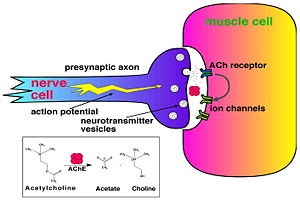
Acetylcholinesterase (AChE) is key enzyme in the nervous system of animals. By rapid hydrolysis of the neurotransmitter, acetylcholine (ACh), AChE terminates neurotransmission at cholinergic synapses. It is a very fast enzyme, especially for a serine hydrolase, functioning at a rate approaching that of a diffusion-controlled reaction. AChE inhibitors are among the key drugs approved by the FDA for management of Alzheimer's disease (AD). The powerful toxicity of organophosphorus (OP) poisons is attributed primarily to their potent AChE inhibitors.
Key Enzyme in the Nervous System
|
Solution of the three-dimensional (3D) structure of Torpedo californica acetylcholinesterase (TcAChE) in 1991 opened up new horizons in research on an enzyme that had already been the subject of intensive investigation.[1] The unanticipated structure of this extremely rapid enzyme, in which the active site was found to be buried at the bottom of a , lined by (colored dark magenta), led to a revision of the views then held concerning substrate traffic, recognition and hydrolysis.[2] To understand how those aromatic residues behave with the enzyme, see Flexibility of aromatic residues in acetylcholinesterase. Solution of the 3D structure of acetylcholinesterase led to a series of theoretical and experimental studies, which took advantage of recent advances in theoretical techniques for treatment of proteins, such as molecular dynamics and electrostatics and to site-directed mutagenesis, utilizing suitable expression systems. Acetylcholinesterase hydrolysizes the neurotransmitter acetylcholine , producing group. directly binds (via its nucleophilic Oγ atom) within the catalytic triad (ACh/TcAChE structure 2ace). The residues are also important in the ligand recognition.
Treatment of Alzheimer's disease
Alzheimer's disease (AD) is a disorder that attacks the central nervous system through progressive degeneration of its neurons. AD occurs in around 10% of the elderly and, as yet, there is no known cure. Patients with this disease develop dementia which becomes more severe as the disease progresses. It was suggested that symptoms of AD are caused by decrease of activity of cholinergic neocortical and hippocampal neurons. Treatment of AD by ACh precursors and cholinergic agonists was ineffective or caused severe side effects. ACh hydrolysis by AChE causes termination of cholinergic neurotransmission. Therefore, compounds which inhibit AChE might significantly increase the levels of ACh depleted in AD. Indeed, it was shown that AChE inhibitors improve the cognitive abilities of AD patients at early stages of the disease development. The first generation of AD drugs were AChE inhibitors: alcaloids like (-)-Huperzine A (HupA) and (-)-galanthamine (GAL, Reminyl); synthetic compounds tacrine (Cognex) and rivastigmine (Exelon).
(-)-Huperzine A, discovered by Chinese scientists from 1980s, has been proved to be a powerful, highly specific, and reversible inhibitor of AChE. It is a novel alkaloid originally isolated from the Traditional Chinese medicine [1] Qian Ceng Ta which is produced from the whole plant of the firmossHuperzia serrata. Qian Ceng Ta has been used for over 1000 years in China for treatment of contusions, strains, swellings, schizophrenia and myasthenia gravis. Shuangyiping[2], a tablet form of HupA produced from the extracts of Huperzia serrata, was developed in 1996 as a new drug for symptomatic treatment of Alzheimer’s disease in China. Compared with the other three FDA-approved drugs for the treatment of Alzheimer’s disease, Donepezil (Aricept), Rivastigmine (Exelon), Galanthamine (Reminyl), HupA has better penetration through the blood-brain barrier, higher oral bioavailability, and longer duration of AChE inhibitory action. The structure of HupA shows some similarity to other known AChE inhibitors. The molecule is fairly rigid and contains an aromatic system as well as a primary amino group that is probably protonated at physiological pH. Various suggestions have been made with respect to its orientation within the active site of AChE, and with respect to the amino acid residue with which its putative pharmacophoric groups might interact. Solution of the 3D structure of a complex of HupA with AChE would permit unequivocal resolution of this issue and it would also provide a rational basis for structure-related drug design aimed at developing synthetic analogues of HupA with improved therapeutic properties.
|
The crystal structure of the complex of TcAChE with HupA at 2.5 Å resolution (1vot) was determined in 1997 and it shows an unexpected orientation for the inhibitor with surprisingly few strong direct interactions with protein residues to explain its high affinity. HupA binds to TcAChE at the active site, and its in comparison to ACh. The principal interactions of are including: a direct (colored orange) through a water molecule as a linker at the bottom of the gorge; cation-π interactions between the amino group of (colored lime) with the distance between the nitrogen and the centroid of the aromatic rings of 4.8 and 4.7 Å, respectively; at the top of the gorge, hydrogen bonds through two water molecules as linkers formed between the amino group of (colored magenta). An unusually short (~3.0 Å) C-H→O HB has been seen between the ethylidene methyl group of (colored crimson).
|
Galanthamine. (red) is an alkaloid from the flower snowdrop (Galanthus nivalis). The X-ray crystal structure of the TcAChE/GAL complex (1dx6) was determined at 2.3 Å resolution. The inhibitor binds at the base of the active site gorge of TcAChE, interacting with both the choline-binding site (Trp84) and the acyl-binding pocket (Phe288, Phe290). The tertiary amine appears to make a non-conventional hydrogen bond, via its N-methyl group, to Asp72. The hydroxyl group of the inhibitor makes a strong hydrogen bond (2.7 Å) with Glu199. ACh (gray) is shown for comparison.
|
Tacrine. In the X-ray crystal structure of TcAChE/ complex which was determined at 2.8 Å resolution, the tacrine is seen (magenta) bound in the active site of TcAChE (1acj) . ACh (gray) is shown for comparison.
|
Rivastigmine (Exelon) is a carbamate inhibitor of AChE, and it is currenly used in therapy of Alzheimer's disease. Rivastigmine (colored yellow) interacts with TcAChE (colored lime) at the (1gqr). The carbamyl moiety of rivastigmine is to the active-site S200 Oγ. The second part of rivastigmine (the leaving group), NAP ((−)-S-3-[1-(dimethylamino)ethyl]phenol) is also held in the active-site gorge, but it is from the carbamyl moiety, hence, carbamylation took place. The of TcAChE/NAP (colored magenta) is known (1gqs). The TcAChE active-site residues which are interacting with NAP are colored violet. NAP is located in a similar region of TcAChE active site, but with different orientation than that of the NAP part (colored yellow) in the TcAChE/rivastigmine complex. Only H440 and F330 significantly change their side-chain conformations. of the TcAChE active sites in 4 different structures (TcAChE/rivastigmine (1gqr), TcAChE/NAP (1gqs), native TcAChE (2ace), and TcAChE/VX (1vxr, TcAChE colored white and VX black) reveals that the conformation of H440 in the TcAChE/NAP structure is very similar its conformation in the native TcAChE (2ace), but the distance between H440 Nδ and E327 Oε is significantly longer in the TcAChE/rivastigmine and the TcAChE/VX complexes. This structural change disrupts the catalytic triad consisting of S200, E327, H440. This could explain the very slow kinetics of AChE reactivation after its inhibition by rivastigmine.
|
The active site gorge has , a catalytic site (consisting of the catalytic triad together with Trp84 & Phe330) and a peripheral site (including Trp 279 & Tyr 121), which helps prebind the substrate and direct it toward the active site. The 3D structure showed not only that the active site was buried deep in the enzyme, but surprisingly, there were no negatively charged residues along this gorge, as was expected to help attract the positively charged ACh substrate, rather, instead, a series of aromatic residues that are highly conserved in all AChE sequences. See: AChE inhibitors and substrates
Selected 3D Structures of AChE
Acetylcholinesterase - AChE native
3lii – hAChE - recombinant human
1ea5, 2ace – TcAChE – trigonal – Torpedo californica
2j3d – TcAChE – monoclinic
1w75 – TcAChE – orthorhombic
1eea – TcAChE – cubic
2vt6, 2vt7 – TcAChE – different dosage
1qid to 1qim - TcAChE synchrotron radiation damage
1j06, 1maa – mAChE - mouse
1qo9 – DmAChE - Drosophila
1c2o, 1c2b – electrophorus AChE – Electric eel
AChE inhibitors (In Different Languages)
1eve AChE-Aricept complex, 1eve (Arabic), 1eve (Chinese), 1eve (Italian), 1eve (Russian), 1eve (Spanish), 1eve (Turkish)
1vot AChE-Huperzine A complex, 1vot (Chinese)
AChE active site inhibitors conjugating at the bottom of the active site gorge
2w9i – TcAChE + methylene blue
2wls – MosAChE + AMTS13
2vq6 – TcAChE + 2-PAM
2j3q – TcAChE + Thioflavin T
2ha0 – mAChE + ketoamyltrimethylammonium
2h9y – mAChE + TMTFA
1gpk, 1gpn, 1vot – TcAChE + huperzine
1gqr – TcAChE + rivastigmine
1gqs – TcAChE + NAP
1e66 – TcAChE + huprine
1dx4, 1qon – DmAChE + tacrine derivative
1oce – TcAChE + MF268
1ax9, 1ack – TcAChE + edrophonium
1amn – TcAChE + TMTFA
1acj – TcAChE + tacrine
AChE peripheral site inhibitors conjugating at the surface of the protein
1ku6 - mAChE + fasciculin 2
1ku6, 1mah - mAChE + fasciculin 2
1j07 - mAChE + decidium
1n5m - mAChE + gallamine
1n5r - mAChE + propidium
1b41, 1f8u - hAChE + fasciculin 2
1fss - TcAChE + fasciculin 2
AChE bis inhibitors spanning the active site gorge
3i6m – TcAChE + N-piperidinopropyl galanthamine
3i6z - TcAChE + saccharinohexyl galanthamine
1zgb, 1zgc – TcAChE + tacrine (10) hupyridone
2w6c – TcAChE + bis-(-)-nor-meptazinol
2ckm, 2cmf – TcAChE + bis-tacrine
2cek – TcAChE + N-[8-(1,2,3,4-tetrahydroacridin-9-ylthio)octyl]-1,2,3,4-tetrahydroacridin-9-amine
1ut6 - TcAChE + N-9-(1,2,3,4-tetrahydroacridinyl)-1,8-diaminooctane
1odc - TcAChE + N-4-quinolyl-N-9-(1,2,3,4-tetrahydroacridinyl)-1,8-diaminooctane
1w4l, 1w6r, 1w76, 1dx6, 1qti - TcAChE + galanthamine and derivative
1q83, 1q84 - mAChE + TZ2PA6
1h22, 1h23 – TcAChE + bis-hupyridone
1hbj – TcAChE + quinoline derivativev
1e3q – TcAChE + bw284c51
1eve – TcAChE + e2020
1acl – TcAChE + decamethonium
AChE organophosphate inhibitors causing irreversible inhibition
2wu3 – mAChE + fenamiphos and HI-6
2wu4 – mAChE + fenamiphos and ortho-7
2jgf - mAChE + fenamiphos
2wfz, 2wg0, 1som - TcAChE + soman
2wg1 - TcAChE + soman + 2-PAM
2whp, 2whq, 2whr – mAChE + sarin and HI-6
2jgg - mAChE + sarin
2jgl - mAChE + VX and sarin
1cfj - TcAChE + sarin, GB
3dl4, 3dl7 – mAChE + tabun
2jey – mAChE + HLO-7
2c0p, 2c0q - mAChE + tabun
2jez - mAChE + tabun + HLO-7
2jf0 - mAChE + tabun + Ortho-7
2jgh - mAChE + VX
1vxo, 1vxr - TcAChE + VX
2jgi, 2jgm - mAChE + DFP
1dfp - TcAChE + DFP
2jgj, 2jgk, 2jge - mAChE + methamidophos
2gyu - mAChE + HI-6
2gyv - mAChE + Ortho-7
2gyw - mAChE + obidoxime
AChE substrate analogues mimicking the binding of the substrate acetylcholine
2ha4 – mAChE (mutant) + acetylcholine
2vja, 2vjb, 2vjc, 2vjd, 2cf5 – TcAChE + 4-oxo-N,N,N-trimethylpentanaminium
2v96, 2v97, 2v98, 2v99 – TcAChE + 1-(2-nitrophenyl)-2,2,2-trifluoroethyl-arsenocholine
2ha2 – mAChE + succinylcholine
2ha3 - mAChE + choline
2ha5 – mAChE (mutant) + acetylthiocholine
2ha6 – mAChE (mutant) + succinylthiocholine
2ha7 – mAChE (mutant) + butyrylthiocholine
2ch4, 2c58 – TcAChE + acetylthiocholine
2c5g – TcAChE + thiocholine
Others...
2j4f – TcAChE + Hg
1vzj – TcAChE tetramerization domain
1jjb – TcAChE + PEG
Additional Resources
For additional information, see: Alzheimer's Disease
External Links
- Acetylcholinesterase Tutorial by Karl Oberholser, Messiah College
- PDB Molecule of the Month - Acetylcholinesterase
- Movies: X-ray Damage in ACh & Nature's Vacuum Cleaner by R. Gillilan, Cornell Univ
References
- ↑ Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I. Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein. Science. 1991 Aug 23;253(5022):872-9. PMID:1678899
- ↑ Botti SA, Felder CE, Lifson S, Sussman JL, Silman I. A modular treatment of molecular traffic through the active site of cholinesterase. Biophys J. 1999 Nov;77(5):2430-50. PMID:10545346
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Joel L. Sussman, Alexander Berchansky, David Canner, Eran Hodis, Clifford Felder, Jaime Prilusky, Harry Greenblatt, Yechun Xu