We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Shank protein
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
βPIX belongs to a group of guanine nucleotide exchange factors used by Rho GTPase family members, like Rac1 and Cdc42, which are known to regulate the actin cytoskeleton of synapses.<ref name="IM">PMID: 20117114</ref> PIX has an N-terminal Src homology 3 (SH3) domain which associates with PAK, a coiled-coil (CC) domain, which is critical for multimerization, and a C-terminal PDZ binding domain which interacts with the PDZ domain of Shank.<ref name="IM"/> The interaction of Shank with βPIX promotes the synaptic localization of βPIX and βPIX associated p21 Associated Kinase (PAK). Since PAK is known to regulate actin cytoskeletons and dendritic spines are actin-rich structures, it is believed that Shank recruits βPIX and associated proteins to spines to regulate the PSD.<ref name="Park"/> | βPIX belongs to a group of guanine nucleotide exchange factors used by Rho GTPase family members, like Rac1 and Cdc42, which are known to regulate the actin cytoskeleton of synapses.<ref name="IM">PMID: 20117114</ref> PIX has an N-terminal Src homology 3 (SH3) domain which associates with PAK, a coiled-coil (CC) domain, which is critical for multimerization, and a C-terminal PDZ binding domain which interacts with the PDZ domain of Shank.<ref name="IM"/> The interaction of Shank with βPIX promotes the synaptic localization of βPIX and βPIX associated p21 Associated Kinase (PAK). Since PAK is known to regulate actin cytoskeletons and dendritic spines are actin-rich structures, it is believed that Shank recruits βPIX and associated proteins to spines to regulate the PSD.<ref name="Park"/> | ||
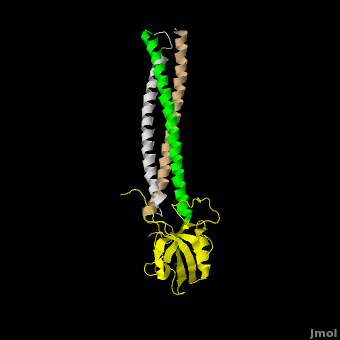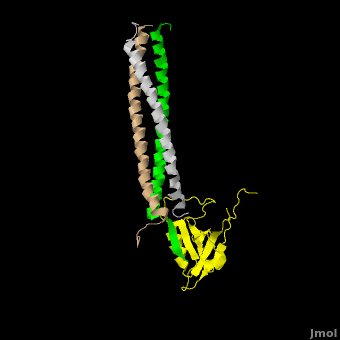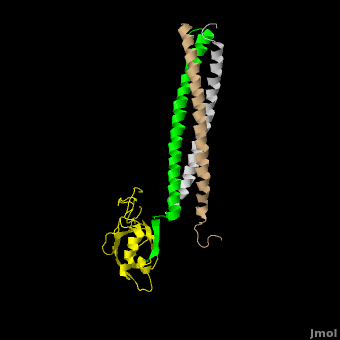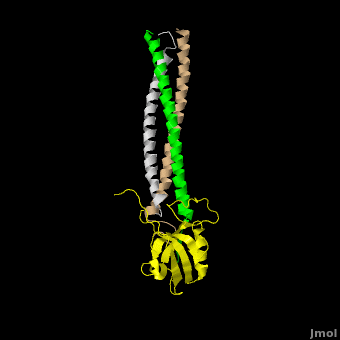
| - | The <scene name='Shank_Family_Proteins/Pdz/1'>canonical PDZ domain</scene> contains 90 amino acids and folds into a compact <scene name='Shank_Family_Proteins/Pdz_glob/1'>globular structure</scene> consisting of a six-stranded β-sandwich flanked by two alpha helices.<ref name="IM"/> βPIX possess a <scene name='Shank_Family_Proteins/Bpix_trimer/2'>parallel trimer</scene> via <scene name='Shank_Family_Proteins/Bpix_phob/1'>helical hydrophobic interactions</scene> within its CC domain, a <scene name='Shank_Family_Proteins/Proline/1'>proline to break the helix</scene>, and a <scene name='Shank_Family_Proteins/Pdz_binding/1'>PDZ binding domain</scene> at the C-terminus. Interestingly, only 1 Shank molecule is bound to the CC domain trimer of βPIX in an | + | The <scene name='Shank_Family_Proteins/Pdz/1'>canonical PDZ domain</scene> contains 90 amino acids and folds into a compact <scene name='Shank_Family_Proteins/Pdz_glob/1'>globular structure</scene> consisting of a six-stranded β-sandwich flanked by two alpha helices.<ref name="IM"/> βPIX possess a <scene name='Shank_Family_Proteins/Bpix_trimer/2'>parallel trimer</scene> via <scene name='Shank_Family_Proteins/Bpix_phob/1'>helical hydrophobic interactions</scene> within its CC domain, a <scene name='Shank_Family_Proteins/Proline/1'>proline to break the helix</scene>, and a <scene name='Shank_Family_Proteins/Pdz_binding/1'>PDZ binding domain</scene> at the C-terminus. Interestingly, only 1 Shank molecule is bound to the CC domain trimer of βPIX in an <scene name='Shank_Family_Proteins/Asym/1'>asymettric assembly</scene>. The <scene name='Shank_Family_Proteins/Bubble/1'>8-residue PDZ binding domain</scene> (BALL AND STICK AND SPHERE COMBO BURIED MODE) of βPIX forms a number of **hydrogen bonding and hydrophobic interactions** (FIGURE 2A) with the Shank PDZ domain. Shank3-Arg 679 forms the **most critical interaction** with βPIX, tightly binding Glutamate -3. Abolishing this interaction through mutagenesis completely eliminates the assembly. Upon binding of βPIX, the PDZ domain undergoes a significant <scene name='Shank_Family_Proteins/Morph_overview/4'>conformational change</scene>. Lys 682 undergoes a nearly <scene name='Shank_Family_Proteins/Morph_lys/3'>11 Angstrom displacement</scene> to make room for the βPIX PDZ binding domain.<ref name="IM"/> |
Shank proteins are positioned between scaffolding proteins that are bound to either neurotransmitter receptors or the actin cytoskeleton. This puts Shank proteins in a perfect position to nucleate the underlying structure of the PSD.<ref name="Baron"/> The SAM domain of <scene name='Shank_Family_Proteins/Multimer_opening/1'>Shank3 can oligomerize</scene> (<scene name='Shank_Family_Proteins/Multimer_opening_alt/2'>Alternate View</scene>) to form large sheets composed of helical fibers stacked side by side. The proposed sheet structure with radially projecting protein interaction domains, is ideal architecture for a protein that must contact both membrane and cytoplasmic components at a synaptic surface.<ref name="Baron"/> Models of this sort validate the importance of Shank3 as master scaffolding proteins and illustrate how slight mutations can disrupt an entire PSD and synaptic function. | Shank proteins are positioned between scaffolding proteins that are bound to either neurotransmitter receptors or the actin cytoskeleton. This puts Shank proteins in a perfect position to nucleate the underlying structure of the PSD.<ref name="Baron"/> The SAM domain of <scene name='Shank_Family_Proteins/Multimer_opening/1'>Shank3 can oligomerize</scene> (<scene name='Shank_Family_Proteins/Multimer_opening_alt/2'>Alternate View</scene>) to form large sheets composed of helical fibers stacked side by side. The proposed sheet structure with radially projecting protein interaction domains, is ideal architecture for a protein that must contact both membrane and cytoplasmic components at a synaptic surface.<ref name="Baron"/> Models of this sort validate the importance of Shank3 as master scaffolding proteins and illustrate how slight mutations can disrupt an entire PSD and synaptic function. | ||
Revision as of 05:09, 3 March 2011
| |||||||||||
Additional Structures of Shank Family Proteins
References
- ↑ 1.0 1.1 1.2 Park E, Na M, Choi J, Kim S, Lee JR, Yoon J, Park D, Sheng M, Kim E. The Shank family of postsynaptic density proteins interacts with and promotes synaptic accumulation of the beta PIX guanine nucleotide exchange factor for Rac1 and Cdc42. J Biol Chem. 2003 May 23;278(21):19220-9. Epub 2003 Mar 7. PMID:12626503 doi:10.1074/jbc.M301052200
- ↑ 2.0 2.1 2.2 Baron MK, Boeckers TM, Vaida B, Faham S, Gingery M, Sawaya MR, Salyer D, Gundelfinger ED, Bowie JU. An architectural framework that may lie at the core of the postsynaptic density. Science. 2006 Jan 27;311(5760):531-5. PMID:16439662 doi:311/5760/531
- ↑ 3.0 3.1 3.2 Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsater H, Sponheim E, Goubran-Botros H, Delorme R, Chabane N, Mouren-Simeoni MC, de Mas P, Bieth E, Roge B, Heron D, Burglen L, Gillberg C, Leboyer M, Bourgeron T. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007 Jan;39(1):25-7. Epub 2006 Dec 17. PMID:17173049 doi:ng1933
- ↑ 4.0 4.1 4.2 4.3 Bozdagi O, Sakurai T, Papapetrou D, Wang X, Dickstein DL, Takahashi N, Kajiwara Y, Yang M, Katz AM, Scattoni ML, Harris MJ, Saxena R, Silverman JL, Crawley JN, Zhou Q, Hof PR, Buxbaum JD. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol Autism. 2010 Dec 17;1(1):15. PMID:21167025 doi:10.1186/2040-2392-1-15
- ↑ Garber K. Neuroscience. Autism's cause may reside in abnormalities at the synapse. Science. 2007 Jul 13;317(5835):190-1. PMID:17626859 doi:10.1126/science.317.5835.190
- ↑ Abu-Elneel K, Liu T, Gazzaniga FS, Nishimura Y, Wall DP, Geschwind DH, Lao K, Kosik KS. Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetics. 2008 Jul;9(3):153-61. Epub 2008 Jun 19. PMID:18563458 doi:10.1007/s10048-008-0133-5
- ↑ 7.0 7.1 7.2 7.3 Im YJ, Kang GB, Lee JH, Park KR, Song HE, Kim E, Song WK, Park D, Eom SH. Structural basis for asymmetric association of the betaPIX coiled coil and shank PDZ. J Mol Biol. 2010 Mar 26;397(2):457-66. Epub 2010 Jan 29. PMID:20117114 doi:10.1016/j.jmb.2010.01.048
Proteopedia Page Contributors and Editors (what is this?)
David Canner, Michal Harel, Alexander Berchansky, Joel L. Sussman