User:Kai D. Ludwig/Sandbox 1
From Proteopedia
| Line 6: | Line 6: | ||
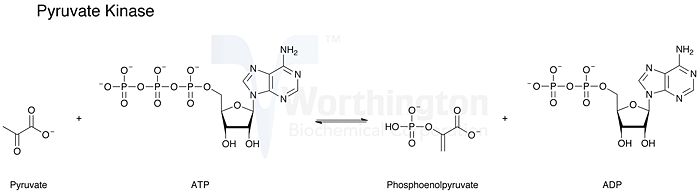
[[Image:Pk_reaction.jpg|700px|right|thumb|Reaction Catalyzed by Pyruvate Kinase, | [[Image:Pk_reaction.jpg|700px|right|thumb|Reaction Catalyzed by Pyruvate Kinase, | ||
image taken from http://www.worthington-biochem.com/PKL/images/reaction.jpg]] | image taken from http://www.worthington-biochem.com/PKL/images/reaction.jpg]] | ||
| - | Pyruvate Kinase is | + | Pyruvate Kinase(PK) is the enzyme in the final step of aerobic glycolysis. The enzyme catalyzes the transfer of a phosphate group from phosphoenolpyruvate (PEP) to an adenosine diphosphate (ADP) to make both pyruvate and adenosine triphosphate (ATP)1. The reaction is shown below in figure 1. The purpose for a cell to perform glycolysis is to create energy in the form of ATP as well as to make pyruvate. Pyruvate is then converted and used to create even more energy in the cell. Pyruvate can either be directed to enter the Krebs cycle when in aerobic conditions or else be made into lactate. The conversion to lactate produces more ATP and occurs in aerobic conditions, or lacking oxygen. It has also been shown that UTP, GTP, CTP, ITP, and dATP can act as donors2. This reaction has a large negative free energy that results from the substrate level phosphorylation (making of ATP). The negative or Gibbs free energy that results from breaking a phosphate group off of phosphoenolpyruvate is 12.3 kcal/mol or 51.6 kJ/mol. Being such a significantly large release of energy, the reaction is essentially reversible. This allows the process to be a regulator in the glycolytic process. Pyruvate Kinase is regulated by its own substrates. Increased regulation occurs when PEP is high in concentration and contrarily, PK is inhibited in the presence of its products: ATP and pyruvate. It makes sense that pyruvate kinase would increase its catabolic activities in the presence of more PEP so it can make more energy (ATP) and create pyruvate. This is true except if the cell already contains large amount of ATP and pyruvate, hence they inhibit pyruvate kinase. |
[[Image:Pyruvate Kinase Mol of the Month(glycolytic enzymes).gif|300px|left|thumb|Pyruvate Kinase, | [[Image:Pyruvate Kinase Mol of the Month(glycolytic enzymes).gif|300px|left|thumb|Pyruvate Kinase, | ||
image taken from http://www.pdb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/pdb50_11.html 'Molecule of the Month' Glycolytic Enzymes:Pyruvate Kinase]] | image taken from http://www.pdb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/pdb50_11.html 'Molecule of the Month' Glycolytic Enzymes:Pyruvate Kinase]] | ||
| Line 12: | Line 12: | ||
---- | ---- | ||
| - | === | + | ===ISOZYMES=== |
| - | + | The human has several different forms of pyruvate kinase. The two different isozymes are the L/R and M form. They are isozymes because the two forms different greatly in there amino acid sequence but have the exact same function. The isozymes are tissue specific and are named accordingly: L (liver), R (erythrocyte), and M (Muscle). The muscle form can be either considered M1 or M2. M1 is found primarily in muscle, heart and brain and the M2 form is found in fast proliferating cells such as early fetal tissue and cancers as well as in the kidney.** The canonical form or primary form considered in literature is the R-type of pyruvate kinase. The R-type found in red blood cells is very similar to the L-type found in hepatocytes and differ only in the sequence of the first 33 amino acids. They sequence difference is as follows: MSIQENISSLQLRSWVSKSQRDLAKSILIGAPG (R)→ ME (L).** This sequence difference may lead to a difference in allosteric regulation properties. The M1 and M2 form have a major difference in their amino acid sequence from the R/L form. Yet, they have the same catalyitic activity. The M1 difference from the M2 type in its sequence from amino acids 389-433 as is as follows: IYHLQLFEELRRLAPITSDPTEATAVGAVEASFKCCSGAIIVLTK (M2)→ MFHRKLFEELVRASSHSTDL MEAMAMGSVEASYKCLAAAL IVLTE (M1). ** This sequence difference leads to binding activities and regulation of its activity that are discussed later. | |
---- | ---- | ||
| - | === | + | ===STRUCTURE=== |
| - | + | Pyruvate Kinase in its active form is a homotetramer. This means it consists of four identical subunits or monomers. Each monomer also consists of four different domains. They are the N-terminal, A Domain, B Domain, and C-terminal. Many parts of pyruvate kinase are arranged into secondary structures for stability. Pyruvate kinase contains both alpha helixes as well as beta sheets. Each subunit of pyruvate kinase contains all four domains as well as an active site. This different “pieces” of pyruvate kinase do not facilitate a reaction as individual parts. Instead they come together to form the homotetramer or quaternary structure. In addition to needing all four subunits, cofactors are necessary to function. Each monomer requires Mg2+ (or Mn2+) and K+. These cofactors act on specific amino acids.** This allows for the enzyme to change conformation or overall shape. When bound, pyruvate kinase changes from its low affinity T state to its high substrate affinity R state. This action can be described as the enzyme ‘breathing’ as it contracts and expands in the presence of co-factors. Other co-factors include Fructose 1,6-bisphosphate (FBP) and 2-phosphoglycolate (PGA). | |
---- | ---- | ||
Revision as of 15:22, 14 April 2011
Contents |
PYRUVATE KINASE
INTRODUCTION
Pyruvate Kinase(PK) is the enzyme in the final step of aerobic glycolysis. The enzyme catalyzes the transfer of a phosphate group from phosphoenolpyruvate (PEP) to an adenosine diphosphate (ADP) to make both pyruvate and adenosine triphosphate (ATP)1. The reaction is shown below in figure 1. The purpose for a cell to perform glycolysis is to create energy in the form of ATP as well as to make pyruvate. Pyruvate is then converted and used to create even more energy in the cell. Pyruvate can either be directed to enter the Krebs cycle when in aerobic conditions or else be made into lactate. The conversion to lactate produces more ATP and occurs in aerobic conditions, or lacking oxygen. It has also been shown that UTP, GTP, CTP, ITP, and dATP can act as donors2. This reaction has a large negative free energy that results from the substrate level phosphorylation (making of ATP). The negative or Gibbs free energy that results from breaking a phosphate group off of phosphoenolpyruvate is 12.3 kcal/mol or 51.6 kJ/mol. Being such a significantly large release of energy, the reaction is essentially reversible. This allows the process to be a regulator in the glycolytic process. Pyruvate Kinase is regulated by its own substrates. Increased regulation occurs when PEP is high in concentration and contrarily, PK is inhibited in the presence of its products: ATP and pyruvate. It makes sense that pyruvate kinase would increase its catabolic activities in the presence of more PEP so it can make more energy (ATP) and create pyruvate. This is true except if the cell already contains large amount of ATP and pyruvate, hence they inhibit pyruvate kinase.

ISOZYMES
The human has several different forms of pyruvate kinase. The two different isozymes are the L/R and M form. They are isozymes because the two forms different greatly in there amino acid sequence but have the exact same function. The isozymes are tissue specific and are named accordingly: L (liver), R (erythrocyte), and M (Muscle). The muscle form can be either considered M1 or M2. M1 is found primarily in muscle, heart and brain and the M2 form is found in fast proliferating cells such as early fetal tissue and cancers as well as in the kidney.** The canonical form or primary form considered in literature is the R-type of pyruvate kinase. The R-type found in red blood cells is very similar to the L-type found in hepatocytes and differ only in the sequence of the first 33 amino acids. They sequence difference is as follows: MSIQENISSLQLRSWVSKSQRDLAKSILIGAPG (R)→ ME (L).** This sequence difference may lead to a difference in allosteric regulation properties. The M1 and M2 form have a major difference in their amino acid sequence from the R/L form. Yet, they have the same catalyitic activity. The M1 difference from the M2 type in its sequence from amino acids 389-433 as is as follows: IYHLQLFEELRRLAPITSDPTEATAVGAVEASFKCCSGAIIVLTK (M2)→ MFHRKLFEELVRASSHSTDL MEAMAMGSVEASYKCLAAAL IVLTE (M1). ** This sequence difference leads to binding activities and regulation of its activity that are discussed later.
STRUCTURE
Pyruvate Kinase in its active form is a homotetramer. This means it consists of four identical subunits or monomers. Each monomer also consists of four different domains. They are the N-terminal, A Domain, B Domain, and C-terminal. Many parts of pyruvate kinase are arranged into secondary structures for stability. Pyruvate kinase contains both alpha helixes as well as beta sheets. Each subunit of pyruvate kinase contains all four domains as well as an active site. This different “pieces” of pyruvate kinase do not facilitate a reaction as individual parts. Instead they come together to form the homotetramer or quaternary structure. In addition to needing all four subunits, cofactors are necessary to function. Each monomer requires Mg2+ (or Mn2+) and K+. These cofactors act on specific amino acids.** This allows for the enzyme to change conformation or overall shape. When bound, pyruvate kinase changes from its low affinity T state to its high substrate affinity R state. This action can be described as the enzyme ‘breathing’ as it contracts and expands in the presence of co-factors. Other co-factors include Fructose 1,6-bisphosphate (FBP) and 2-phosphoglycolate (PGA).
TUMOR PKM2
|