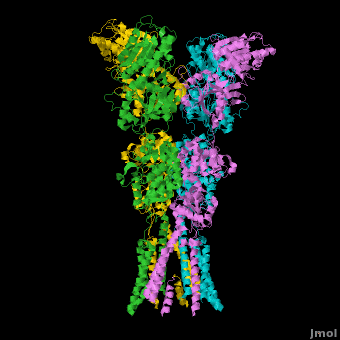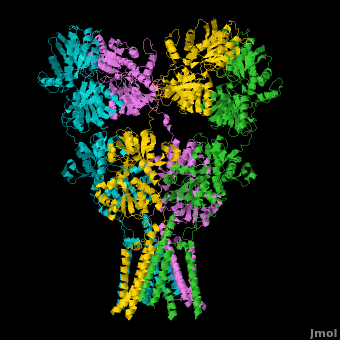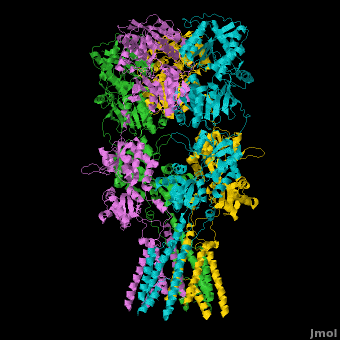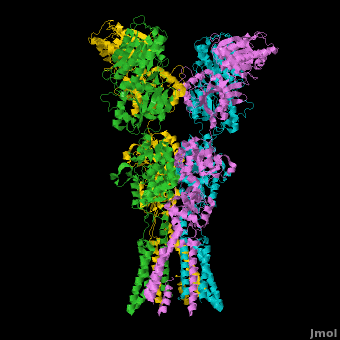Ionotropic Glutamate Receptors
From Proteopedia
| Line 1: | Line 1: | ||
<StructureSection load='' size='500' side='right' caption='Structure of the Ionotropic Glutamate Receptor, GluA2, ([[3kg2]])' scene='Ionotropic_Glutamate_Receptors/Opening/1'> | <StructureSection load='' size='500' side='right' caption='Structure of the Ionotropic Glutamate Receptor, GluA2, ([[3kg2]])' scene='Ionotropic_Glutamate_Receptors/Opening/1'> | ||
[[Image:IGluR Picture.png|200px|left]] [[Ionotropic Glutamate Receptors]] '''(IGluRs)''' are a family of ligand-gated ion channels that are responsible for fast excitatory neurotransmission.<ref name="Jin">PMID: 16192394</ref> Primarily localized to nerve synapses in mammals, IGluRs are implicated in nearly all aspects of nervous system development and function.<ref name="Sobo">PMID: 19946266</ref> Synapses form the connection between two neuronal cells. Within synapses, neurotransmitters are released from vesicles in presynaptic cells and interact with receptors in postsynaptic cells to allow for communication between nerve cells.<ref name="Jin"/> Glutamate is the predominant neurotransmitter of excitatory synapses and interacts specifically with AMPA and NMDA IGluRs.<ref name="Purcel"/> | [[Image:IGluR Picture.png|200px|left]] [[Ionotropic Glutamate Receptors]] '''(IGluRs)''' are a family of ligand-gated ion channels that are responsible for fast excitatory neurotransmission.<ref name="Jin">PMID: 16192394</ref> Primarily localized to nerve synapses in mammals, IGluRs are implicated in nearly all aspects of nervous system development and function.<ref name="Sobo">PMID: 19946266</ref> Synapses form the connection between two neuronal cells. Within synapses, neurotransmitters are released from vesicles in presynaptic cells and interact with receptors in postsynaptic cells to allow for communication between nerve cells.<ref name="Jin"/> Glutamate is the predominant neurotransmitter of excitatory synapses and interacts specifically with AMPA and NMDA IGluRs.<ref name="Purcel"/> | ||
| + | See details of ionotropic glutamate receptor 2 i [[Molecular Playground/Glutamate Receptor]]. | ||
====Involvement in Autism Spectrum Disorders==== | ====Involvement in Autism Spectrum Disorders==== | ||
[[Autism Spectrum Disorders]] (ASDs) are neurodevelopmental disorders. During development, glutamate regulates neuronal growth and synaptogenesis, effectively dictating the underlying connective neuronal architecture of the brain.<ref name="Purcel"/> Significant research into ASDs has been devoted to understanding how glutamate receptors function and how disruption leads to neurodevelopmetnal disorders. IGluRs are concentrated in regions of the brain that have been implicated in ASDs including the cerebellum and hippocampus. These are the areas responsible for motor control, spatial navigation and memory, attributes that are often disrupted in patients with ASDs. Studies have revealed that glutamate receptor expression is increased in the cerebellum of autistic individuals by nearly 250%. A number of small nucleotide polymorphisms in IGluRs have also been identified which correlate with the ASDs. Further, many people with autism have clearly visible disturbances in the inferior olive (IO) in the brain. The IO plays a critical role in movement coordination and maintenance of an underlying 12 Hz brain rhythm through careful regulation of glutamate signaling.<ref>PMID: 15749250</ref> A well-known mouse model called “Lurcher” for the lurching type movements the mice make has served as an important model for studying ASDs. The mutation that causes the “Lurcher” phenotype creates a constitutively leaky glutamate receptor ion channel resulting in IO neuron degeneration and loss of purkinje cells. These mice exhibit some of the well-known Autism-like characteristics.<ref>PMID: 9285588</ref> Such relationships between overly active glutamate receptors leading to increased excitation/inhibition ratios and autism have led some to propose using glutamate receptor inhibitors as a means of [[Pharmaceutical Drugs|pharmaceutical intervention]] for improving those with autistic symptoms.<ref>PMID: 14606691</ref> Many pharmacological agents that reduce neural excitation, such as benzodiazapines, are thought to potentially have therapeutic value in treating autistic symptoms.<ref name="Purcel">PMID: 11706102</ref> | [[Autism Spectrum Disorders]] (ASDs) are neurodevelopmental disorders. During development, glutamate regulates neuronal growth and synaptogenesis, effectively dictating the underlying connective neuronal architecture of the brain.<ref name="Purcel"/> Significant research into ASDs has been devoted to understanding how glutamate receptors function and how disruption leads to neurodevelopmetnal disorders. IGluRs are concentrated in regions of the brain that have been implicated in ASDs including the cerebellum and hippocampus. These are the areas responsible for motor control, spatial navigation and memory, attributes that are often disrupted in patients with ASDs. Studies have revealed that glutamate receptor expression is increased in the cerebellum of autistic individuals by nearly 250%. A number of small nucleotide polymorphisms in IGluRs have also been identified which correlate with the ASDs. Further, many people with autism have clearly visible disturbances in the inferior olive (IO) in the brain. The IO plays a critical role in movement coordination and maintenance of an underlying 12 Hz brain rhythm through careful regulation of glutamate signaling.<ref>PMID: 15749250</ref> A well-known mouse model called “Lurcher” for the lurching type movements the mice make has served as an important model for studying ASDs. The mutation that causes the “Lurcher” phenotype creates a constitutively leaky glutamate receptor ion channel resulting in IO neuron degeneration and loss of purkinje cells. These mice exhibit some of the well-known Autism-like characteristics.<ref>PMID: 9285588</ref> Such relationships between overly active glutamate receptors leading to increased excitation/inhibition ratios and autism have led some to propose using glutamate receptor inhibitors as a means of [[Pharmaceutical Drugs|pharmaceutical intervention]] for improving those with autistic symptoms.<ref>PMID: 14606691</ref> Many pharmacological agents that reduce neural excitation, such as benzodiazapines, are thought to potentially have therapeutic value in treating autistic symptoms.<ref name="Purcel">PMID: 11706102</ref> | ||
Revision as of 09:39, 25 July 2011
| |||||||||||
Page Development
This article was developed based on lectures given in Chemistry 543 by Prof. Clarence E. Schutt at Princeton University.
3D structures of glutamate receptor
Ionotropic glutamate receptor 0
2pyy - IGluR2 ligand-binding domain + Glu – Nostoc punctiforme
Ionotropic glutamate receptor 2
3n6v, 3o2j - rIGluR2 N-terminal domain (mutant) – rat
3hsy, 3h5v, 3h5w - rIGluR2 N-terminal domain
2wjw, 2wjx – hIGluR2 N-terminal domain - human
3rn8, 3rnn – hIGluR2 ligand-binding domain + potentiator
3bki - rIGluR2 ligand-binding domain + inhibitor
1fto - rIGluR2 ligand-binding domain
GluR2 positive allosteric modulator complex
2xhd - hIGluR2 ligand-binding domain + positive allosteric modulator + Glu
2al4, 1p1o - rIGluR2 ligand-binding domain (mutant) + positive allosteric modulator
2xx8, 2xx7, 2xx9, 2xxh, 2xxi, 3lsf, 3h6t, 3h6u, 3h6v, 3h6w, 3bbr - rIGluR2 ligand-binding domain (mutant) + positive allosteric modulator + Glu
3pmv, 3pmw, 3pmx, 3o28, 3o29, 3o2a, 3o6g, 3o6h, 3o6i, 3m3l, 3lsl - rIGluR2 ligand-binding domain + positive allosteric modulator + Glu
1mm6, 1mm7 - rIGluR2 ligand-binding domain + positive allosteric modulator
3ijo, 3ijx, 3ik6, 3il1, 3ilt, 3ilu - rIGluR2 + positive allosteric modulator + Glu
GluR2 antagonist complex
3r7x - hIGluR2 + antagonist
3kgc, 3kg2, 2cmo - rIGluR2 ligand-binding domain + antagonist + Glu
3h03, 3h06, 3b7d, 1n0t, 1ftl - rIGluR2 ligand-binding domain + antagonist
1lb9 - rIGluR2 ligand-binding domain (mutant) + antagonist
GluR2 agonist complex
3rtf, 3rtw, 3pd8, 3pd9, 3bft, 3bfu, 1wvj, 1syh, 1syi, 1ms7, 1mqd, 1nnp, 1nnk, 1m5b, 1m5c, 1m5d, 1m5e, 1m5f, 1ftm - rIGluR2 ligand-binding domain + agonist
3b6t, 2al5, 2anj, 1p1q, 1p1u, 1p1w, 1lb8 - rIGluR2 ligand-binding domain (mutant) + agonist
1lbc - rIGluR2 ligand-binding domain (mutant) + agonist + Glu
2p2a - rIGluR2 ligand-binding domain + agonist + Glu
GluR2 partial agonist complex
1y1m, 2aix, 1y1z, 1y20, 1mqg, 1mqh, 1mqi, 1mqj, 1mxu, 1mxv, 1mxw, 1mxx, 1mxy, 1mxz, 1my0, 1my1, 1my2, 1my3, 1my4, 1fw0, 1ftk, 1gr2 - rIGluR2 ligand-binding domain + partial agonist
1xhy, 1p1n, 1lbb - rIGluR2 ligand-binding domain (mutant) + partial agonist
GluR2 ligand complex
3dp6, 2uxa, 2i3v, 2i3w, 1ftj - rIGluR2 ligand-binding domain + Glu
3b6q, 3b6w, 2gfe - rIGluR2 ligand-binding domain (mutant) + Glu
Ionotropic glutamate receptor 3
3o21, 3p3w – rIGluR3 N-terminal domain
3m3k – rIGluR3 ligand-binding domain
3rt6, 3rt8, 3dp4 – rIGluR3 ligand-binding domain + agonist
3m3f – rIGluR3 ligand-binding domain + allosteric modulator
3dln - rIGluR3 ligand-binding domain + Glu
Ionotropic glutamate receptor 4
3epe, 3fas - rIGluR4 ligand-binding domain + Glu
3kei – rIGluR4 ligand-binding domain (mutant) + Glu
3kfm - rIGluR4 ligand-binding domain (mutant) + partial agonist
3en3 - rIGluR4 ligand-binding domain + partial agonist
3fat – rIGluR4 ligand-binding domain + agonist
Ionotropic glutamate receptor 5
3fuz, 2zns – hIGluR5 ligand-binding domain + Glu
1txf - rIGluR5 ligand-binding domain + Glu
2ojt - rIGluR5 + anion
3fv1, 3fv2, 3fvg, 3fvk, 3fvn, 3fvo, 2znt, 2znu - hIGluR5 ligand-binding domain + agonist
3c31, 3c32, 3c33, 3c34, 3c35, 3c36 - rIGluR5 ligand-binding domain + ion
2wky, 3gba, 3gbb, 2pbw, 2f34, 2f35, 2f36 – rIGluR5 ligand-binding domain + agonist
2qs1, 2qs2, 2qs3, 2qs4 - rIGluR5 ligand-binding domain (mutant) + agonist
1vso - rIGluR5 ligand-binding domain + antagonist
Ionotropic glutamate receptor 6
3h6g, 3h6h – rIGluR6 N-terminal domain
3g3f, 1sd3, 1s50, 1s7y – rIGluR6 ligand-binding domain + Glu
3g3g, 3g3h, 3g3i, 3g3j, 3g3k, 2i0b, 2i0c - rIGluR6 ligand-binding domain (mutant) + Glu
1s9t – rIGluR6 ligand-binding domain + positive allosteric modulator
1tt1 – rIGluR6 ligand-binding domain + partial agonist
Metabotropic glutamate receptor 1
1ewt, 1ewv - rMGluR1 ligand-binding domain
3ks9 – hMGluR1 (mutant) + antagonist
1isr, 1ewk – rMGluR1 ligand-binding domain + Glu
1iss - rMGluR1 ligand-binding domain + antagonist
Metabotropic glutamate receptor 3
2e4u – hMGluR3 ligand-binding domain (mutant) + Glu
2e4v, 2e4w, 2e4x, 2e4y, 2e4z - hMGluR3 ligand-binding domain (mutant) + agonist
Metabotropic glutamate receptor 5
3lmk – hMGluR5 ligand-binding domain (mutant) + positive allosteric modulator + Glu
Metabotropic glutamate receptor 7
3mq4 – hMGluR7 ligand-binding domain + antagonist
Ionotropic kainate receptor 1
1ycj – rGluK1 ligand-binding domain + Glu
Ionotropic kainate receptor 2
2xxr – rGluK2 ligand-binding domain + Glu
2xxu, 2xxx – rGluK2 ligand-binding domain (mutant) + Glu
2xxt – rGluK2 ligand-binding domain + partial agonist
2xxv, 2xxy – rGluK2 ligand-binding domain (mutant) + partial agonist
1yae - rGluK2 ligand-binding domain + agonist
Ionotropic kainate receptor 3
3olz – rGluK3 N-terminal domain
Ionotropic kainate receptor 5
3om0, 3om1 – rGluK5 N-terminal domain
NMDA receptor
3jpy, 3jpw – rNMDA subunit ε2 N-terminal domain (mutant)
2a5t - rNMDA subunits NR1 and NR2A ligand-binding domains
2a5s - rNMDA subunits NR1 and NR2A ligand-binding domains + Glu
Additional Resources
For additional information on the Symmetry of the Glutamate Receptor, See: Glutamate Receptor Symmetry Analysis
For Additional Information, See: Membrane Channels & Pumps
For Additional Information, See: Alzheimer's Disease
References
- ↑ 1.0 1.1 1.2 Jin R, Clark S, Weeks AM, Dudman JT, Gouaux E, Partin KM. Mechanism of positive allosteric modulators acting on AMPA receptors. J Neurosci. 2005 Sep 28;25(39):9027-36. PMID:16192394 doi:25/39/9027
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009 Dec 10;462(7274):745-56. Epub . PMID:19946266 doi:10.1038/nature08624
- ↑ 3.0 3.1 3.2 3.3 Purcell AE, Jeon OH, Zimmerman AW, Blue ME, Pevsner J. Postmortem brain abnormalities of the glutamate neurotransmitter system in autism. Neurology. 2001 Nov 13;57(9):1618-28. PMID:11706102
- ↑ Welsh JP, Ahn ES, Placantonakis DG. Is autism due to brain desynchronization? Int J Dev Neurosci. 2005 Apr-May;23(2-3):253-63. PMID:15749250 doi:10.1016/j.ijdevneu.2004.09.002
- ↑ Zuo J, De Jager PL, Takahashi KA, Jiang W, Linden DJ, Heintz N. Neurodegeneration in Lurcher mice caused by mutation in delta2 glutamate receptor gene. Nature. 1997 Aug 21;388(6644):769-73. PMID:9285588 doi:10.1038/42009
- ↑ Rubenstein JL, Merzenich MM. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2003 Oct;2(5):255-67. PMID:14606691
- ↑ Jin R, Singh SK, Gu S, Furukawa H, Sobolevsky AI, Zhou J, Jin Y, Gouaux E. Crystal structure and association behaviour of the GluR2 amino-terminal domain. EMBO J. 2009 Jun 17;28(12):1812-23. Epub 2009 May 21. PMID:19461580 doi:10.1038/emboj.2009.140
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, David Canner, Wayne Decatur, Alexander Berchansky, Joel L. Sussman