We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Mati
From Proteopedia
(Difference between revisions)
(→structure) |
|||
| Line 16: | Line 16: | ||
<scene name='Sandbox/Jil/1'>active site</scene> | <scene name='Sandbox/Jil/1'>active site</scene> | ||
| + | |||
| + | ==Structure== | ||
| + | <StructureSection load='1dq8' size='500' side='left' scene='HMG-CoA_Reductase/1dq8_starting_scene/1' caption='Crystal Structure of HMG-CoA, [[1dq8]])'> | ||
| + | ===General Structure=== | ||
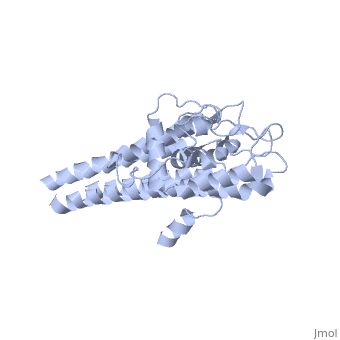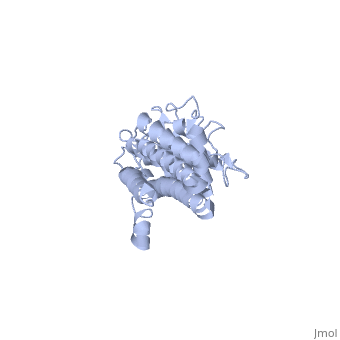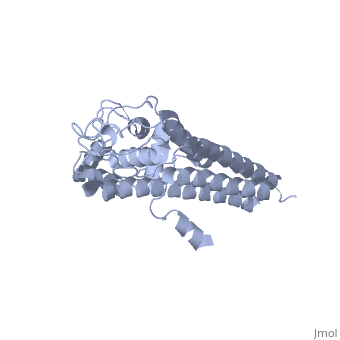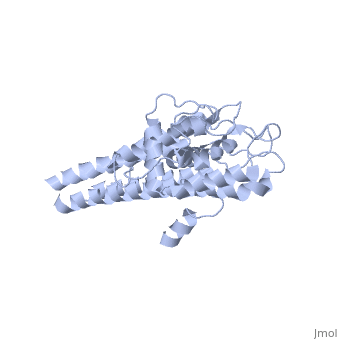
| + | There are two distinct classes of HMGRs, class I HMGRs, which are only found in eukaryotes and are membrane bound and class II HMGRs, which are found in prokaryotes and are soluble.<ref>PMID:11349148</ref> HMGR contains 8 transmembrane domains that have yet to be successfully crystallized, which anchor the protein to the membrane of the endoplasmic reticulum.<ref name="Roitelman"/> The catalytic portion of human HMGR forms a tetramer, with the individual monomers winding around each other.<ref name="Roitelman">PMID:1374417</ref> Within the tetramer, the monomers are arranged into <scene name='HMG-CoA_Reductase/1dq8_2_dimers/3'>two dimers</scene>, each of which contains <scene name='HMG-CoA_Reductase/1dq8_2_active_sites/2'>two active sites </scene>which are formed by residues form both monomers. Each monomer contains <scene name='HMG-CoA_Reductase/1dq8_star3_domains/2'>three domains </scene>, the <scene name='HMG-CoA_Reductase/1dq8_n_domain/2'>N-domain</scene>, the <scene name='HMG-CoA_Reductase/1dq8_l_domain/1'>L-Domain</scene>, and the <scene name='HMG-CoA_Reductase/1dq8_s_domain/1'>S-Domain</scene>. The L-domain is unique to HMGRs while the S-domain, which forms the binding site for NADP, resembles that of [[ferredoxin]]. The S and L domains are connected by a <scene name='HMG-CoA_Reductase/1dq8_cis_loop/5'>“cis-loop”</scene> which is essential for the HMG-binding site.<ref name="Roitelman"/> Salt bridges between residues R641 and E782 as well as <scene name='HMG-CoA_Reductase/1dq8_cis_loop/4'>hydrogen bonds</scene> between E700 and E700 on neighboring monomers compliment the largely hydrophobic dimer-dimer interface.<ref name="Roitelman"/> | ||
| + | <br /> | ||
Revision as of 18:40, 23 April 2012
This Sandbox page is available for temporary practice work. Nothing in a Sandbox page is permanent. You may prefer to create your own personal Sandbox page -- see instructions. Feel free to add practice content below this paragraph, or delete everything below this paragraph, but please do not delete this paragraph.
|
Contents |
structure
more structure
Level 3
Level 4
Level 5
| |||||||||
| 1l8w, resolution 2.30Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Non-Standard Residues: | |||||||||
| |||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Image:Hammock in the sea.jpg
Caption
Structure
| |||||||||||