Human SUMO E1~SUMO1-AMP tetrahedral intermediate mimic
From Proteopedia
| Line 1: | Line 1: | ||
| - | <StructureSection load='3kyd' size='400' frame='true' | + | <StructureSection load='3kyd' size='400' frame='true' side='right' scene='3kyd/Cv/1' > |
[[Image:3kyd.png|left|200px]] | [[Image:3kyd.png|left|200px]] | ||
===Human SUMO E1~SUMO1-AMP tetrahedral intermediate mimic ([[3kyd]])=== | ===Human SUMO E1~SUMO1-AMP tetrahedral intermediate mimic ([[3kyd]])=== | ||
Current revision
| |||||||||||
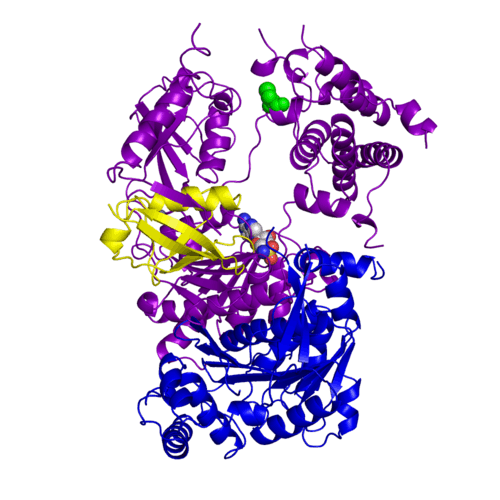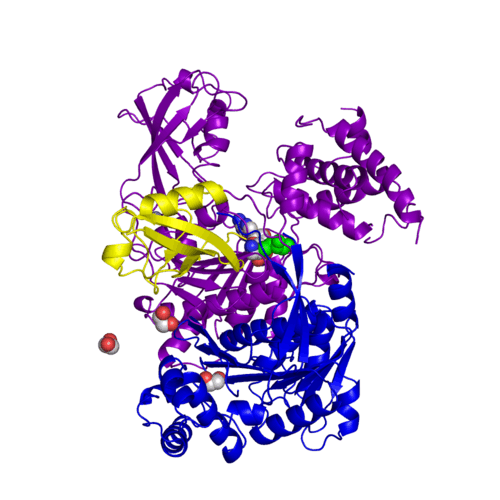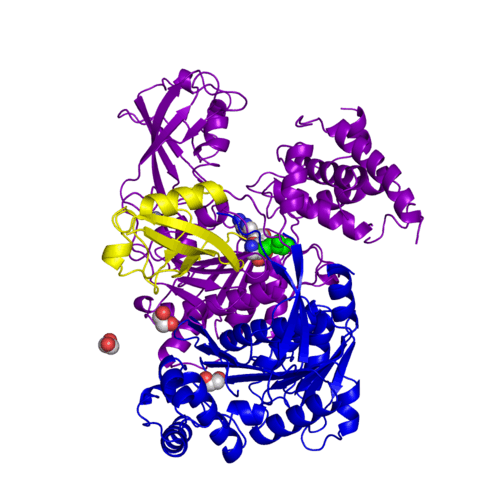
For better understanding of the difference between these two conformations you can see this morph (generated by using POLYVIEW-3D: http://polyview.cchmc.org/polyview3d.html; reload/refresh this page to restart this movie). Of note, in contrast to the previous figure, the same domains of these two structures (3kyc and 3kyd) are colored in the same colors (SUMO1 in yellow, SAE1 colored in blue and other domains in darkviolet). The catalytic Cys173 is shown in the spacefill representation and colored green, AMSN (or AVSN) are shown in the spacefill representation and colored in CPK colors.
About this Structure
3KYD is a 3 chains structure with sequences from Homo sapiens. Full crystallographic information is available from OCA.
Reference
- Olsen SK, Capili AD, Lu X, Tan DS, Lima CD. Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature. 2010 Feb 18;463(7283):906-12. PMID:20164921 doi:10.1038/nature08765
Categories: Homo sapiens | Lima, C D. | Acetylation | Adenylation | Atp-binding | Cytoplasm | E1 | Inhibitor | Isopeptide bond | Ligase | Membrane | Nucleotide-binding | Nucleus | Phosphoprotein | Polymorphism | Sumo | Tetrahedral intermediate | Thioester | Ubiquitin | Ubl conjugation pathway