User:Erin May/Sandbox 1
From Proteopedia
| Line 60: | Line 60: | ||
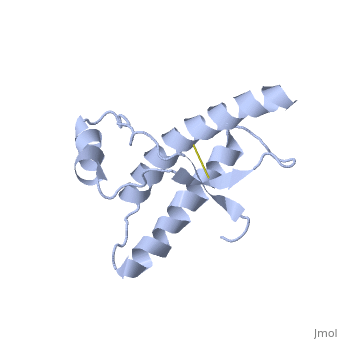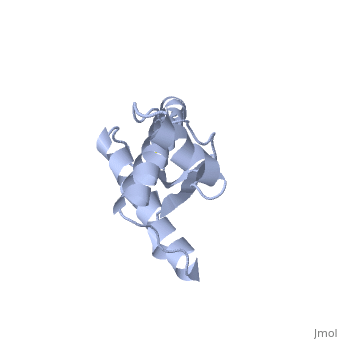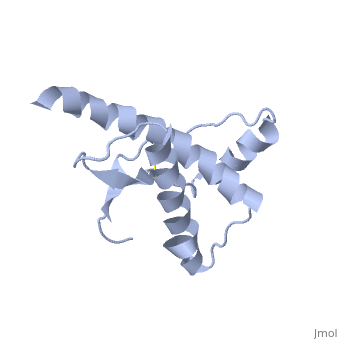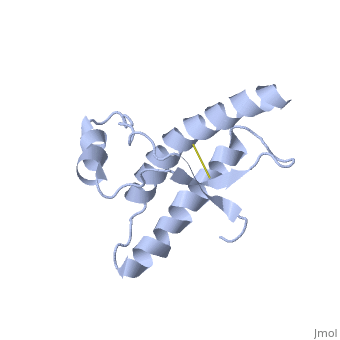
The specific residues shown alter the electrostatic properties of the surface of PrP<sup>C</sup>, however their postions on the wild-type monomer and fully unfolded PrP<sup>Sc</sup>, do not illustrate a clear mechanism for propagation. The dimer form clearly shows the reason for the infectious qualities of the variable and disease conforming residues. | The specific residues shown alter the electrostatic properties of the surface of PrP<sup>C</sup>, however their postions on the wild-type monomer and fully unfolded PrP<sup>Sc</sup>, do not illustrate a clear mechanism for propagation. The dimer form clearly shows the reason for the infectious qualities of the variable and disease conforming residues. | ||
| - | It is theorized from this dimeric structure that the dimerization is the first step in amyloid formation. | + | It is theorized from this dimeric structure that the dimerization is the first step in amyloid formation and the presence of these dimers could possibly speed up the aggregation of PrP<sup>Sc</sup>. |
Residues 129, 200, and 164 to 170 are shown to exist right at the dimer interfaces. | Residues 129, 200, and 164 to 170 are shown to exist right at the dimer interfaces. | ||
Helix 1 (Ser 143−Tyr 157) exists at the dimer interface. <ref name="Knaus">PMID:11524679</ref> | Helix 1 (Ser 143−Tyr 157) exists at the dimer interface. <ref name="Knaus">PMID:11524679</ref> | ||
| + | Helix 2 (Asn 171−Thr 188) is linked to the C-terminal helix 3 (Thr 199−Tyr 225). | ||
| + | The switch region 189-198 is shown to be unfolded. | ||
| + | There are also additional intrachain van der Waals and electrostatic interactions between the switch region and helix 1 in the 3D domain-swapped dimeric crystal structure that are not possible in the monomeric NMR structure. | ||
| + | |||
| + | Hydrogen bonding between the dimers exist: Thr188 O−Gly195 N, Thr190 O−Lys194 N and Thr192 O−Thr192 N | ||
| + | |||
| + | Altered in familial: 119, 129, 226 shown | ||
| + | |||
| + | Asp 202 and Arg 220, which are located at either end of helix 3, form interchain hydrogen bonds that confer specificity to the interaction | ||
| + | |||
| + | Due to the importance of initial dimerization in the formation of prion aggregates, the step of dimerization presents a known step for treatments to target. | ||
</StructureSection> | </StructureSection> | ||
Revision as of 07:04, 27 November 2012
Contents |
Prions as a disease causing agent
Prions are infectious or genetic misfolded proteins which act as templates upon which properly folded prion protein monomers can aggregate. Prions contain no nucleic acid such as other infectoius molecules or organisms. Human Prion Protein or Major Prion protein, exists as a normal constituent of human cells, found mostly in the brain[2] and is called PrPC.[3] PrPC is composed of mostly helix whereas the infectious form, PrPSc, is composed of high percentage beta sheets.[3]
The diseases prions confer are neurodegenerative disorders which result from the large scale aggregation of these proteins. For more information about the infections related to prions see Transmissible spongiform encephalopathy at Wikipedia.
Unfolding Mechanism
Currently, the mechanism by which a template prion unfolds a the helices of a properly folded prion protein is unknown. Specific residues have been shown to either confer resistance or lend themselves to this unfolding.
PrPC natural monomer
| |||||||||||
PrPSc
| |||||||||||
Dimer Form
| |||||||||||
Reference List
- ↑ Image of Creutzfeldt-Jakob positive brain tissue was obtained from The CDC's Public Health Image Library.
- ↑ Centers for Disease Control and Prevention
- ↑ 3.0 3.1 Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998 Nov 10;95(23):13363-83. PMID:9811807
- ↑ 4.0 4.1 Lee S, Antony L, Hartmann R, Knaus KJ, Surewicz K, Surewicz WK, Yee VC. Conformational diversity in prion protein variants influences intermolecular beta-sheet formation. EMBO J. 2010 Jan 6;29(1):251-62. Epub 2009 Nov 19. PMID:19927125 doi:10.1038/emboj.2009.333
- ↑ 5.0 5.1 5.2 Zhang Y, Swietnicki W, Zagorski MG, Surewicz WK, Sonnichsen FD. Solution structure of the E200K variant of human prion protein. Implications for the mechanism of pathogenesis in familial prion diseases. J Biol Chem. 2000 Oct 27;275(43):33650-4. PMID:10954699 doi:10.1074/jbc.C000483200
- ↑ 6.0 6.1 6.2 Knaus KJ, Morillas M, Swietnicki W, Malone M, Surewicz WK, Yee VC. Crystal structure of the human prion protein reveals a mechanism for oligomerization. Nat Struct Biol. 2001 Sep;8(9):770-4. PMID:11524679 doi:10.1038/nsb0901-770