Sandbox 719
From Proteopedia
| Line 1: | Line 1: | ||
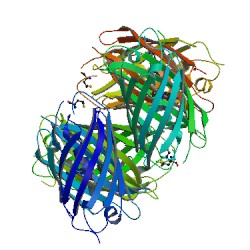
<Structure load='1G7K' size='500' frame='true' align='left' '3D structure of the DsRed, a red fluorescent protein from coral (1G7K) ''Cristalized withe a resolution of 2.0 Å'''/> | <Structure load='1G7K' size='500' frame='true' align='left' '3D structure of the DsRed, a red fluorescent protein from coral (1G7K) ''Cristalized withe a resolution of 2.0 Å'''/> | ||
In this article, we want to present you a red fluorescent Protein recently cloned from a corallimorpharian of the ''Discosoma'' genus. The PDB number of this molecule is 1G7K <ref>http://www.rcsb.org/pdb/explore/explore.do?structureId=1G7K</ref> | In this article, we want to present you a red fluorescent Protein recently cloned from a corallimorpharian of the ''Discosoma'' genus. The PDB number of this molecule is 1G7K <ref>http://www.rcsb.org/pdb/explore/explore.do?structureId=1G7K</ref> | ||
| - | The publications related is called "Refined crystal structure of DsRed, a red fluorescent protein from carol, at 2.0-Å resolution" <ref>Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0-A resolution. | + | The publications related is called "Refined crystal structure of DsRed, a red fluorescent protein from carol, at 2.0-Å resolution" <ref>Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0-A resolution. Yarbrough D, Wachter RM, Kallio K, Matz MV, Remington SJ. PubMed : [http://www.ncbi.nlm.nih.gov/pubmed/11209050 11209050] |
| - | Yarbrough D, Wachter RM, Kallio K, Matz MV, Remington SJ. | + | PubMedCentral : [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC14609/ PMC14609] DOI : [http://www.pnas.org/content/98/2/462 10.1073]</ref> |
| - | PubMed : [http://www.ncbi.nlm.nih.gov/pubmed/11209050 11209050] | + | |
| - | PubMedCentral : [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC14609/ PMC14609] | + | |
| - | DOI : [http://www.pnas.org/content/98/2/462 10.1073]<ref | + | |
| - | + | ||
| - | + | ||
| - | + | ||
== The DsRed, a red fluorescent protein from coral == | == The DsRed, a red fluorescent protein from coral == | ||
Revision as of 18:48, 6 January 2013
|
In this article, we want to present you a red fluorescent Protein recently cloned from a corallimorpharian of the Discosoma genus. The PDB number of this molecule is 1G7K [1] The publications related is called "Refined crystal structure of DsRed, a red fluorescent protein from carol, at 2.0-Å resolution" [2]
Contents |
The DsRed, a red fluorescent protein from coral
Introduction
Structure of the DsRed protein
Structure Determination
Primary Structure
Secondary Structure
The tetramer Interface
- Active Site
- Ligands
Structure around the Chromophore
Obtaining of this structure
Mutation studies
Several studies were lead on DsRed, by random and directed mutagenesis. These studies allowed to identify the influence of a certain number of amino acids on characteristics or on the 3D structur of the protein, and to understand the mechanism of the fluorophore. The replacement of Lys-70 by a Met (mutation K70M) lead to a green fluorescent protein, very close to GFP. The reason of this modification is that Lys residue interacts directly with the chromophore. In fact Lys-70 is certainly a charged residue and should interacts via a water molecule with the residues Glu-215 and Glu-148 of the chromophore. The replacement by the neutral residue Met make this interaction impossible. Mutation of Asn-42 (which should interacts with Gln-66) can be understood by the same way, and lead also to a green molecule. An other interesting mutation is the replacement of Lys-83. The effect of this mutation depend of the amino will replace the Lys. Actually, K83R and K83N mutations results in green fluorescent protein, but K83M substitution lead to latge redshift of absorbtion and emission maxima. In fact, substition with an Arg will increased the chain size, and so inhibite the maturation, but substitution with a Met (wich had mainly the same size than Lys, but different charge) change only absorbtion and emission behavior.
Analisys and comparison between DsRed and GFP chromophore
These experiments shown the existence of a green fluorescent protein intermediate, very close to GFP, and suggests that there are several steps in the overall reaction (Matz et al.). Studies of Baird et al. shown there are two key conserved amino acids : Gln-66 and Gln-215. When we do the comparison of chromophore of DsRed and GFP, two points seems to be important. First Gln-66 of DsRed had a sp2 hybridization while Thr/Ser-65 (which are at the same place in the molecule) of GFP have a sp3 hybridization. Secondly, the conserved glutamate Glu-215 of DsRed is closer of the chromophore and is bonded to a water molecule close to GlN-66, which becomes oxidized. So this positionning of Gln-66 and Glu-215 should have a big influence in the red fluorescence, and Glu-215 could have two roles : In the formation of green fluorescence intermediate, and, by its environment, is crucial for formation of green or red emitting species. Matz et al. Suggested that GFP is a “broken” version of an ancestral red fluorescent protein, due to these obsevations.