Hiv-1 gag
From Proteopedia
(Difference between revisions)
| Line 20: | Line 20: | ||
==Nucleocapsid (NC)== | ==Nucleocapsid (NC)== | ||
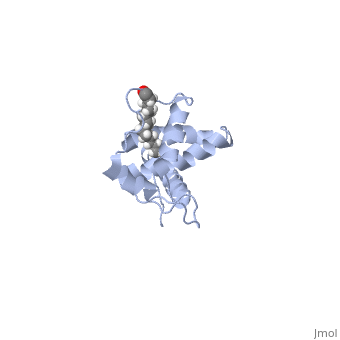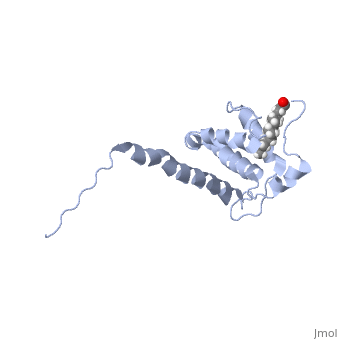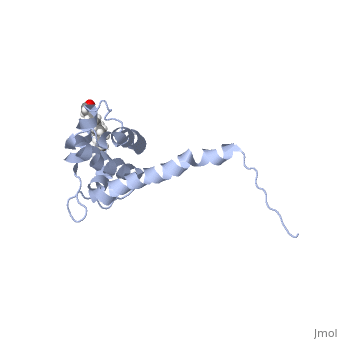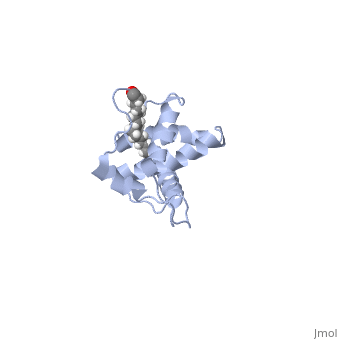
| - | As we have seen, Gag is responsible for correct targeting of viral assembly to discreet sites on the plasma membrane, and viral capsid structure assembly into organized virus particles. However, Gag is also responsible for packaging of the viral RNA into budding virions, and this function is executed by the NC domain. The 5' LTR of HIV-1 genomic RNA contains a recognition element called the psi element. All retroviruses contain some type of psi element in order to get specific packaging of viral genomic RNA within the budding particles, and in the case of HIV-1, the psi element is 120 bases, and contains 4 stem-loop structures. Although the psi element can be quite variable, the SL3 loop is highly conserved within HIV-1 strains. <scene name='Hiv-1_gag/Cv/ | + | As we have seen, Gag is responsible for correct targeting of viral assembly to discreet sites on the plasma membrane, and viral capsid structure assembly into organized virus particles. However, Gag is also responsible for packaging of the viral RNA into budding virions, and this function is executed by the NC domain. The 5' LTR of HIV-1 genomic RNA contains a recognition element called the psi element. All retroviruses contain some type of psi element in order to get specific packaging of viral genomic RNA within the budding particles, and in the case of HIV-1, the psi element is 120 bases, and contains 4 stem-loop structures. Although the psi element can be quite variable, the SL3 loop is highly conserved within HIV-1 strains. <scene name='Hiv-1_gag/Cv/3'>The NC domain of HIV-1 NL4-3 in complex with the SL3 loop of the viral psi element is shown </scene>. |
There are two zinc knuckle domains, with the zinc ion held by three Cys residues and a His (<scene name='User:Nathan_Roy/Zinc_knuckle/1'>Knuckle</scene>). Notice that guanine210 and guanine212 of the RNA interact with the F2 and F1 CCHC zinc knuckles respectively. In the F1 knuckle, G9 fits into a hydrophobic cleft formed by Val13, Phe16, Ile24, and Ala 25. G9 satisfies Watson-Crick hydrogen bonding by interacting with the NH groups from Phe16 and Ala25 (<scene name='User:Nathan_Roy/F1_interaction/1'>Show residues</scene>), and also the CO group of Lys14 (<scene name='User:Nathan_Roy/F1_interaction/2'>Show</scene>). G7 interacts much the same with the F2 knuckle by hydrogen bonding with the NH groups of Trp37 and Met46, while also hydrogen bonding with the CO group of Gly35. The adenine211 nucleotide forms a hydrogen bond with a highly conserved Arg32 residue (<scene name='User:Nathan_Roy/F1_interaction/3'>Show</scene>). Also, residues 3 thru 10 form a 3.10 helix, which nestles into the RNA major groove (<scene name='User:Nathan_Roy/3-10_helix/1'>Show helix</scene>). These interactions allow viral RNA to be specifically packaged into virions. Also of note, it has been shown that NC binding of viral RNA increases the ability of Gag to multimerize, thus providing another mechanism to couple functional virion assembly to Gag multimerization and viral budding. | There are two zinc knuckle domains, with the zinc ion held by three Cys residues and a His (<scene name='User:Nathan_Roy/Zinc_knuckle/1'>Knuckle</scene>). Notice that guanine210 and guanine212 of the RNA interact with the F2 and F1 CCHC zinc knuckles respectively. In the F1 knuckle, G9 fits into a hydrophobic cleft formed by Val13, Phe16, Ile24, and Ala 25. G9 satisfies Watson-Crick hydrogen bonding by interacting with the NH groups from Phe16 and Ala25 (<scene name='User:Nathan_Roy/F1_interaction/1'>Show residues</scene>), and also the CO group of Lys14 (<scene name='User:Nathan_Roy/F1_interaction/2'>Show</scene>). G7 interacts much the same with the F2 knuckle by hydrogen bonding with the NH groups of Trp37 and Met46, while also hydrogen bonding with the CO group of Gly35. The adenine211 nucleotide forms a hydrogen bond with a highly conserved Arg32 residue (<scene name='User:Nathan_Roy/F1_interaction/3'>Show</scene>). Also, residues 3 thru 10 form a 3.10 helix, which nestles into the RNA major groove (<scene name='User:Nathan_Roy/3-10_helix/1'>Show helix</scene>). These interactions allow viral RNA to be specifically packaged into virions. Also of note, it has been shown that NC binding of viral RNA increases the ability of Gag to multimerize, thus providing another mechanism to couple functional virion assembly to Gag multimerization and viral budding. | ||
Revision as of 10:25, 4 April 2013
| |||||||||||
References
- De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998 Jan 16;279(5349):384-8. PMID:9430589
- Ivanov D, Tsodikov OV, Kasanov J, Ellenberger T, Wagner G, Collins T. Domain-swapped dimerization of the HIV-1 capsid C-terminal domain. Proc Natl Acad Sci U S A. 2007 Mar 13;104(11):4353-8. Epub 2007 Mar 5. PMID:17360528
- Gamble TR, Yoo S, Vajdos FF, von Schwedler UK, Worthylake DK, Wang H, McCutcheon JP, Sundquist WI, Hill CP. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997 Oct 31;278(5339):849-53. PMID:9346481
- Ganser-Pornillos BK, Yeager M, Sundquist WI. The structural biology of HIV assembly. Curr Opin Struct Biol. 2008 Apr;18(2):203-17. Epub 2008 Apr 9. PMID:18406133 doi:10.1016/j.sbi.2008.02.001
- Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci U S A. 2006 Jul 25;103(30):11364-9. Epub 2006 Jul 13. PMID:16840558
- Alfadhli A, Barklis RL, Barklis E. HIV-1 matrix organizes as a hexamer of trimers on membranes containing phosphatidylinositol-(4,5)-bisphosphate. Virology. 2009 May 10;387(2):466-72. Epub 2009 Mar 27. PMID:19327811 doi:10.1016/j.virol.2009.02.048
- Jouvenet N, Bieniasz PD, Simon SM. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature. 2008 Jul 10;454(7201):236-40. Epub 2008 May 25. PMID:18500329 doi:10.1038/nature06998