Ciprofloxacin is a broad-spectrum synthetic fluoroquinolone antibiotic that is generally effective against both aerobic gram-positive and aerobic gram-negative bacteria[1].
Bacterial organisms that have been shown to be efficiently targeted by ciprofloxacin are:
Aerobic Gram-Positive Bacteria (With Certain Strain Particularities):
Enterococcus faecalis (many strains are only moderately susceptible), Staphylococcus aureus (methicillin-susceptible strains only), Staphylococcus epidermidis (methicillin-susceptible strains only), Staphylococcus saprophyticus, Streptococcus pneumoniae (penicillin-susceptible strains only), Streptococcus pyogenes.
Aerobic Gram-Negative Bacteria:
Campylobacter jejuni, Citrobacter diversus, Citrobacter freundii, Enterobacter cloacae, Escherichia coli, Haemophilus influenzae, Haemophilus parainfluenzae, Klebsiella pneumoniae, Moraxella catarrhalis, Morganella morganii, Neisseria gonorrhoeae, Proteus mirabilis, Proteus vulgaris, Providencia rettgeri, Providencia stuartii, Pseudomonas aeruginosa, Salmonella typhi, Serratia marcescens, Shigella boydii, Shigella dysenteriae, Shigella flexneri, Shigella sonnei.
Ciprofloxacin also exhibits in vitro minimum inhibitory concentrations of 1 μg/mL or less against strains of the following bacteria (with less adequate characterizations of the effects of treatment against these bacteria in terms of efficiency and general safety):
Aerobic Gram-Positive Bacteria (With Certain Strain Particularities):
Staphylococcus haemolyticus, Staphylococcus hominis, Streptococcus pneumoniae (penicillin-resistant strains only).
Aerobic Gram-Negative Bacteria:
Acinetobacter Iwoffi, Aeromonas hydrophila, Edwardsiella tarda, Enterobacter aerogenes, Klebsiella oxytoca, Vibrio cholerae, Legionella pneumophila, Vibrio parahaemolyticus, Pasteurella multocida, Vibrio vulnificus, Salmonella enteritidis, Yersinia enterocolitica.
Most anaerobic bacteria exhibit Ciprofloxacin-resistance.
The effectiveness of Ciprofloxacin against the anthrax-causing bacteria, Bacillus anthracis - both in vitro and by use of surrogate marker serum levels - has also been demonstrated[2][3]. Thus, Ciprofloxacin is currently a Food and Drug Administration (FDA)-approved treatment for patients who have been exposed to anthrax via inhalation[4]. Likewise, Ciprofloxacin may be used to treat plague (from the bacteria, Yersinia pestis) and tularemia (from the bacteria, Francisella tularensis)[5]. Thus, Ciprofloxacin demonstrates usefulness in the field of counter-bioterrorism given its action against bacteria that could potentially be implemented in biological warfare. Furthermore, in its extended-release tablet form, Ciprofloxacin tends to target certain types of urological infectious agents (e.g. those causing epididymitis). The nature of Ciprofloxacin, then, as a powerful, broad-range antibiotic is crucial for broad-range bacterial infection treatment. An understanding of the action of Ciprofloxacin at the molecular level is, no doubt, necessary for an appreciation of the potency of Ciprofloxacin witnessed at the macro level.
Historical Information
The patented introduction of Ciprofloxacin in the United States occurred in 1987 as a result of the research efforts of Bayer Pharmaceuticals, although there have been reports that at least two European patents had pre-dated the Bayer patent by at least five years[6]. On October 27, 1987, the FDA had approved the drug for use in the United States for the treatment of certain bacterial infections. The effectiveness of Ciprofloxacin as an antibiotic went unchallenged by all alternative antibiotics[7]. Thus, other pharmaceutical companies were forced to offer their alternative antibiotics at lower costs (compared to the cost of Ciprofloxacin) so as to engage any sort of competition with Ciprofloxacin. Because of the tendency of doctors to prescribe lower-cost medication, Bayer Pharmaceuticals could not expand into the international pharmaceutical industry (which, as a whole, was steadily declining) and, consequently, was forced to downsize at the turn of the century. Indeed, the competitive effectiveness of Ciprofloxacin did not overcome the competitive pricing of drugs released by alternative pharmaceutical companies. Faced with the impending expiration of its patent for Ciprofloxacin in the early years of the millennium, Bayer Pharmaceuticals attempted to release variations of Ciprofloxacin. The release of Ciprofloxacin variations such as Pediatric Ciprofloxacin and Once-daily Ciprofloxacin allowed for the extension of the Bayer Pharmaceutical Ciprofloxacin patent. The popularity of Ciprofloxacin rose sharply after September 11, 2001 due its characteristic targeting of anthrax, which was projected as a possible tool for bioterrorism. The prescription of Ciprofloxacin for treatment of bacterial infections continues to this day.
Structure and Administration
General Quinolone-Fluoroquinolone Structure
The identification of Ciprofloxacin as a "quinolone" is a result of the heterocyclic (due to the presence of an inner-ring Nitrogen), bicyclic core-containing structure of Ciprofloxacin, which structure is characteristic of all quinolones[8]. Ciprofloxacin is further characterized as a "fluoroquinolone" since it contains a fluorine atom at the R6 position of its bicyclic core[9]. Indeed, all fluoroquinolones contain this R6 fluorine moiety. A general molecular structure for all fluoroquinolones is shown. The R6 fluorine occurs on the left ring of the bicyclic core.
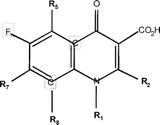 [10]
[10]
Atomic Structure
The specific atomic structure of Ciprofloxacin is shown (all atoms are labeled and numbered).
Administration
Ciprofloxacin is usually administered either as CIPRO® Oral Suspension (Ciprofloxacin) or as CIPRO® Tablets (Ciprofloxacin hydrochloride)[11]. Both administration types are oral.
CIPRO® Oral Suspension (Ciprofloxacin) is a 1-cyclopropyl-6-floro-1,4-dihydro-4-oxo-7-(1-piperazinyl)-3-quinolinecarboxylic acid with empirical formula: C₁₇H₁₈FN₃O₃. Ciprofloxacin has a molecular weight of 331.35 g/mol and occurs as a yellowish, crystalline substance[12][13]. A simple molecular structure of Ciprofloxacin is shown (base empirical formula).
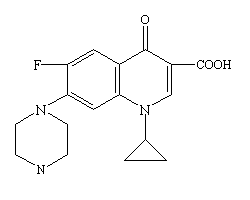 [14]
[14]
CIPRO® Tablet[s] (Ciprofloxacin hydrochloride) is the monohydrochloride monohydrate salt of 1-cyclopropyl-6-floro-1,4-dihydro-4-oxo-7-(1-piperazinyl)-3-quinolinecarboxylic acid with empirical formula C₁₇H₁₈FN₃O₃•HCl•H₂O. Ciprofloxacin hydrochloride has a molecular weight of 385.5 g/mol and also occurs as a yellowish, crystalline substance[15]. A simple molecular structure of Ciprofloxacin hydrochloride is shown.
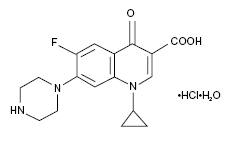 [16]
[16]
Ciprofloxacin may also be administered intravenously and in the form of eye or ear drops[17].
Synthesis
A six-step pathway for Ciprofloxacin synthesis is shown below. This particular pathway is characterized by the initiation of a cyclic chloro-fluoro precursor followed by closing of a nitrogen-containing ring and addition of piperazine ortho with respect to the R6 fluorine. It should be noted, however, that other systems for Ciprofloxacin synthesis have been postulated.
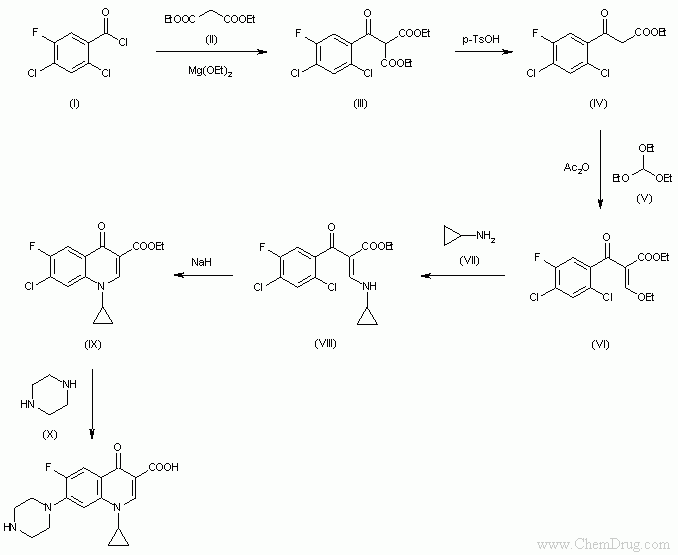 [18]
[18]
Characteristic Protein Targets and Interactions
Ciprofloxacin is known for its efficient ability to hinder bacterial DNA synthesis via inhibition of bacterial DNA Gyrase and DNA Topoisomerase IV. [19]. DNA Gyrase, a type II DNA topoisomerase, is a tetramer composed of 2 GyrA and 2 GyrB subunits. DNA Gyrase is responsible for introducing negative superhelical twists (gyrations, hence, "Gyrase") - as it removes positive superhelical twists - without which twists DNA Replication would not occur. Topoisomerase IV, also a type II DNA topoisomerase, is composed of 2 ParC and 2 ParE subunits, and its overall structure is similar to that of DNA Gyrase. Specifically, ParC is homologous to GyrA, and ParE is homologous to GyrB. Topoisomerase IV is responsible for the separation of interlinked daughter chromosomes, which separation anticipates the segregation of daughter cells. The action of Ciprofloxacin on DNA Gyrase and on Topoisomerase IV is characterized by the stabilization of DNA in complex with either of these two proteins. This stabilization prevents normal motility (and, thus, progression) of the DNA replication fork, which prevention results in a full inhibition of DNA replication. This inhibition ultimately leads to cell death.
DNA Gyrase Target
A twinned structure of is shown. DNA Gyrase is characterized by its "ironing device" appearance with a (in this scene, DNA Gyrase is light blue, and the dip-like cleft runs the length of the base of the protein. The DNA ligand is black and Ciprofloxacin, as in all scenes under this heading, maintains its atomic color labels). Ciprofloxacin intercalates on DNA at (in this scence, DNA is light brown and is in ball-and-stick formation). Ciprofloxacin inhibits the progression of the action of DNA Gyrase on DNA by attacking and stabilizing successive coils of DNA for (in this scene DNA is in mesh formation). The effects of this intercalation on the specific base pairs of participating nucleotides is shown .This intercalation and consequent stabilization prevents proper unwinding of DNA by DNA Gyrase. An example of specifically interrupted sites on the DNA strand is shown . The precise mechanism by which Ciprofloxacin interaction with DNA Gyrase ultimately leads to cell death has not been fully elaborated. However, examination of the location of intercalation of Ciprofloxacin with respect to the amino acid residues of DNA Gyrase near this location leads to the observation that the characteristically polar atoms within the structure of Ciprofloxacin (i.e. fluorine, oxygen, nitrogen) seem to interact with the (in this scene, all polar amino acid residues are blue). A broader analysis of this location indicates that this location, the active site of the protein is composed, primarily, of (in this scene, alpha helices are purple, beta sheets and turns are brown for comparison; DNA is portrayed in dot formation, and Ciprofloxacin is not shown).
Topoisomerase IV Target
The structural characterization of the inhibition of DNA replication via inhibition of the action of DNA Topoisomerase IV by Ciprofloxacin is similar to that via inhibition of the action of DNA Gyrase by Ciprofloxacin. An example structure of is shown (note that this ligand is not Ciprofloxacin, but represents a structure that is analogous to that of Ciprofloxacin). The overall structure of DNA Topoisomerase IV is clearly analogous to that of DNA Gyrase since DNA Topoisomerase IV also appears in an "ironing device" shape with a (in this scene, DNA Topoisomerase IV is light blue, and the dip-like cleft runs the length of the base of the protein. The DNA ligand is black and the example Ciprofloxacin structural analog, as in all scenes under this heading, maintains its atomic color labels). The ligand depicted here intercalates within the DNA structure slightly more aggressively than Ciprofloxacin intercalates within DNA Gyrase (see above), since the DNA structure in this case is (in this scene, DNA is in mesh formation). Yet the concept of obstruction of DNA motility via intercalation applies equivalently in this case and, thus, this model is sufficient for a replication of the action of Ciprofloxacin on DNA within DNA Topoisomerase IV. As expected, based on the aforementioned structural similarities, the interactions between the intercalating ligand (or, Ciprofloxacin) and the active site of DNA Topoisomerase IV are similar to those witnessed between Ciprofloxacin and DNA Gyrase. The active site of the protein is composed, primarily, of (in this scene, alpha helices are purple and polar amino acids on these alpha helices are blue).
Efflux Pump Interaction
Certain bacteria (Escherichia coli, for example) contain a proton motive force-dependent multidrug efflux pump, which, as the name suggests, grants the bacteria resistance to certain drugs [20]. In Escherichia coli, the efflux system that confers particular drug resistance is a tripartite transmembrane resistance structure known as "AcrAB-TolC" [21]. The drug molecule targeted for excretion is captured by the AcrB subunit (most likely from the periplasm or from the periplasm-intermembrane interface) and is then passed on to the TolC complex for final export. Of course, one could argue that the most important member of the AcrAB-TolC resistance complex is the member that is responsible for the initial attraction of the target compound, The AcrB subunit. Ciprofloxacin is one such drug that is for exclusion from the bacterial cell (in this scene, AcrB is in the proposed transmembrane orientation assuming lower cytosolic face and upper exoplasmic face). It has been shown that contribute to the effectiveness of the initial affinity of AcrB for all targets [22] (in this scene, both Phe residues are magenta). It has also been shown that, after ligand binding, a proton may bind to acidic residue in the transmembrane domain, which contains an as yet putative network of electrostatically interacting residues, the perturbation of which interacting residues leads to a series of conformational changes that result in drug expulsion. Residues involved in this chain of events include , and (red, purple, green, respectively). The precise mechanism of the action of the AcrB efflux subunit (and of the tripartite AcrAB-TolC in general) is still under scrutiny.
Conclusion
As indicated in the explanation of the interaction between Ciprofloxacin and DNA Gyrase, the precise mechanisms of all Ciprofloxacin interactions and transport systems have not been fully elaborated. Relevant research, particularly for insight on the precise mechanism for AcrB drug efflux, are currently underway. Regardless of these gaps, it is clear that the action of Ciprofloxacin in vivo is important with respect to the treatment of bacterial infections. Taken from a more global perspective, the action of Ciprofloxacin on protein function seems to indicate a specific field of study that could provide insight into more precise mechanisms for protein function in general. Thus, Ciprofloxacin is indeed a compound of interest in anticipation of a greater understanding of biological functions.
References
- ↑ CIPRO® (ciprofloxacin hydrochloride) TABLETS - CIPRO® (ciprofloxacin*) ORAL SUSPENSION - Drug Information Packet. Bayer HealthCare Pharmaceuticals. Schering Plough Corporation.
- ↑ CIPRO® (ciprofloxacin hydrochloride) TABLETS - CIPRO® (ciprofloxacin*) ORAL SUSPENSION - Drug Information Packet. Bayer HealthCare Pharmaceuticals. Schering Plough Corporation.
- ↑ 2011. Ciprofloxacin. Medicine Plus. American Society of Health-System Pharmacists Inc. 2011. http://www.nlm.nih.gov/medlineplus/druginfo/meds/a688016.html.
- ↑ 2001. Information on Cipro (Ciprofloxacin Hydrochloride) for Inhalation Anthrax for Consumers: Questions and Answers. Fda.gov. http://www.fda.gov/Drugs/EmergencyPreparedness/BioterrorismandDrugPreparedness/ucm130711.htm. Last updated, 2009.
- ↑ 2011. Ciprofloxacin. Medicine Plus. American Society of Health-System Pharmacists Inc. 2011. http://www.nlm.nih.gov/medlineplus/druginfo/meds/a688016.html.
- ↑ Ciprofloxacin. (2010). Pcm.me. http://pcm.me/ciprofloxacin/.
- ↑ Ciprofloxacin - Activity, Business Aspects/Bayer Pharmaceutical. Encyclopedia.jrank.org. http://encyclopedia.jrank.org/articles/pages/1398940/Ciprofloxacin.html
- ↑ Siegmund, K., et al. (2005). Molecular details of quinolone-DNA interactions: solution structure of an unusually stable DNA duplex with covalently linked nalidixic acid residues and non-covalent complexes derived from it. Nucleic Acids [Research], 33(15), 4838-4848.
- ↑ Peterson, L. (2001). Quinolone-Molecular Structure-Activity Relationships: What We Have Learned About Improving Antimicrobial Activity. Clinical Infectious Diseases, 33(3), S180-S186.
- ↑ Image from: http://cid.oxfordjournals.org/content/33/Supplement_3/S180.full.
- ↑ CIPRO® (ciprofloxacin hydrochloride) TABLETS - CIPRO® (ciprofloxacin*) ORAL SUSPENSION - Drug Information Packet. Bayer HealthCare Pharmaceuticals. Schering Plough Corporation.
- ↑ Molecular weight from Chemexper.com.
- ↑ CIPRO® (ciprofloxacin hydrochloride) TABLETS - CIPRO® (ciprofloxacin*) ORAL SUSPENSION - Drug Information Packet. Bayer HealthCare Pharmaceuticals. Schering Plough Corporation.
- ↑ Image from: http://www.google.com/imgres?imgurl=http://textbookofbacteriology.net/themicrobialworld/cipro.gif&imgrefurl=http://textbookofbacteriology.net/themicrobialworld/control.html&usg=__wtzKLHB3NssfnODEB224br5-Bcw=&h=200&w=250&sz=2&hl=en&start=0&zoom=1&tbnid=o7VT7s6FFIUrWM:&tbnh=160&tbnw=199&ei=Hk10TaypBcL58AbyvIjKDw&prev=/images%3Fq%3Dciprofloxacin%26um%3D1%26hl%3Den%26client%3Dfirefox-a%26sa%3DN%26rls%3Dorg.mozilla:en-US:official%26biw%3D1280%26bih%3D647%26tbs%3Disch:1&um=1&itbs=1&iact=hc&vpx=527&vpy=300&dur=1709&hovh=160&hovw=200&tx=155&ty=82&oei=EU10TcvOCMbdtge5msiLDw&page=1&ndsp=16&ved=1t:429,r:7,s:0.
- ↑ Molecular weight from: CIPRO® (ciprofloxacin hydrochloride) TABLETS - CIPRO® (ciprofloxacin*) ORAL SUSPENSION - Drug Information Packet. Bayer HealthCare Pharmaceuticals. Schering Plough Corporation.
- ↑ Image from: http://www.google.com/imgres?imgurl=http://images.rxlist.com/images/rxlist/ciloxan_s.gif&imgrefurl=http://www.rxlist.com/ciloxan_ophthalmic_ointment-drug.htm&usg=__UqTKseSe8hD85c5RLGIz2_dbAg0=&h=142&w=232&sz=2&hl=en&start=16&zoom=1&tbnid=70Q2WG5hppsQ5M:&tbnh=100&tbnw=164&ei=T010TenMFYH_8Aa6gvDKDw&prev=/images%3Fq%3Dciprofloxacin%26um%3D1%26hl%3Den%26client%3Dfirefox-a%26sa%3DN%26rls%3Dorg.mozilla:en-US:official%26biw%3D1280%26bih%3D647%26tbs%3Disch:10%2C624&um=1&itbs=1&iact=hc&vpx=1064&vpy=399&dur=309&hovh=106&hovw=174&tx=98&ty=76&oei=EU10TcvOCMbdtge5msiLDw&page=2&ndsp=18&ved=1t:429,r:17,s:16&biw=1280&bih=647.
- ↑ Ciprofloxacin. (2010). Pcm.me. http://pcm.me/ciprofloxacin/.
- ↑ Image from: http://www.google.com/imgres?imgurl=http://www.chemdrug.com/databases/SYNTHESIS/SYN/09/09000601a.gif&imgrefurl=http://www.chemdrug.com/databases/8_0_dvpytumicutbciwa.html&usg=__TxiDuzCve6C_crxmcPYTpfW5d4s=&h=555&w=678&sz=6&hl=en&start=0&zoom=1&tbnid=xhquLksJBbMnjM:&tbnh=165&tbnw=201&ei=0Y93TdbGI-yI0QGspa25Bw&prev=/images%3Fq%3Dsynthesis%2Bof%2Bciprofloxacin%26um%3D1%26hl%3Den%26client%3Dfirefox-a%26sa%3DN%26rls%3Dorg.mozilla:en-US:official%26biw%3D1280%26bih%3D647%26tbs%3Disch:1&um=1&itbs=1&iact=hc&vpx=346&vpy=105&dur=63&hovh=203&hovw=248&tx=170&ty=128&oei=0Y93TdbGI-yI0QGspa25Bw&page=1&ndsp=16&ved=1t:429,r:1,s:0
- ↑ Ciprofloxacin Oral - Monograph - Ciprofloxacin Hydrochloride. 2009. Medscape.com. http://www.medscape.com/druginfo/monograph cid=med&drugid=7748&drugname=Ciprofloxacin+Oral&monotype=monograph&secid=8.
- ↑ Su, Chih-Chia, et al. (2006). Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway. Journal of Bacteriology, 188(20), 7290-7296.
- ↑ Husain, F., Nikaido, H. (2010). Substrate path in the AcrB multidrug efflux pump of Escherichia coli. Molecular Microbiology, 78(2), 320-330.
- ↑ Su, Chih-Chia, et al. (2006). Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway. Journal of Bacteriology, 188(20), 7290-7296.




