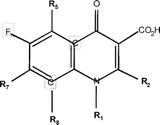
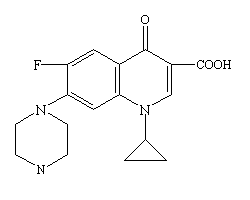
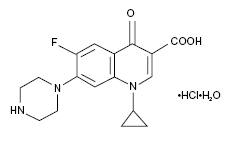
Ciprofloxacin
From Proteopedia
(Difference between revisions)
| Line 77: | Line 77: | ||
=== Efflux Pump Interaction === | === Efflux Pump Interaction === | ||
| - | Certain bacteria (''Escherichia coli'', for example) contain a proton motive force-dependent multidrug <scene name='Ciprofloxacin/Cv/1'>efflux pump</scene>, which, as the name suggests, grants the bacteria resistance to certain drugs <ref>Su, Chih-Chia, et al. (2006). Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway. ''Journal of Bacteriology, 188(20)'', 7290-7296. </ref>. In ''Escherichia coli'', the efflux system that confers particular drug resistance is a tripartite transmembrane resistance structure known as "AcrAB-TolC" <ref>Husain, F., Nikaido, H. (2010). Substrate path in the AcrB multidrug efflux pump of Escherichia coli. ''Molecular Microbiology, 78(2)'', 320-330. </ref>. The drug molecule targeted for excretion is captured by the AcrB subunit (most likely from the periplasm or from the periplasm-intermembrane interface) and is then passed on to the TolC complex for final export. Of course, one could argue that the most important member of the AcrAB-TolC resistance complex is the member that is responsible for the initial attraction of the target compound, the AcrB subunit. Ciprofloxacin may be <scene name='Sandbox_100/Orientation_of_cipro_on_acrb/1'>captured by the AcrB subunit</scene> for removal from the bacterial cell (in this scene, AcrB is in the proposed transmembrane orientation assuming lower cytosolic face and upper exoplasmic face). It has been shown that <scene name='Sandbox_100/Phe_residues/1'> Phe 386 and Phe 388</scene> contribute to the effectiveness of the initial affinity of AcrB for all targets <ref>Su, Chih-Chia, et al. (2006). Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway. ''Journal of Bacteriology, 188(20)'', 7290-7296. </ref> (in this scene, both Phe residues are magenta). It has also been shown that, after ligand binding, a proton may bind to acidic residue in the transmembrane domain, which contains an as yet putative network of electrostatically interacting residues, the perturbation of which interacting residues leads to a series of conformational changes that result in drug expulsion. | + | Certain bacteria (''Escherichia coli'', for example) contain a proton motive force-dependent multidrug <scene name='Ciprofloxacin/Cv/1'>efflux pump</scene>, which, as the name suggests, grants the bacteria resistance to certain drugs <ref>Su, Chih-Chia, et al. (2006). Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway. ''Journal of Bacteriology, 188(20)'', 7290-7296. </ref>. In ''Escherichia coli'', the efflux system that confers particular drug resistance is a tripartite transmembrane resistance structure known as "AcrAB-TolC" <ref>Husain, F., Nikaido, H. (2010). Substrate path in the AcrB multidrug efflux pump of Escherichia coli. ''Molecular Microbiology, 78(2)'', 320-330. </ref>. The drug molecule targeted for excretion is captured by the AcrB subunit (most likely from the periplasm or from the periplasm-intermembrane interface) and is then passed on to the TolC complex for final export. Of course, one could argue that the most important member of the AcrAB-TolC resistance complex is the member that is responsible for the initial attraction of the target compound, the AcrB subunit. Ciprofloxacin may be <scene name='Sandbox_100/Orientation_of_cipro_on_acrb/1'>captured by the AcrB subunit</scene> for removal from the bacterial cell (in this scene, AcrB is in the proposed transmembrane orientation assuming lower cytosolic face and upper exoplasmic face). It has been shown that <scene name='Sandbox_100/Phe_residues/1'> Phe 386 and Phe 388</scene> contribute to the effectiveness of the initial affinity of AcrB for all targets <ref>Su, Chih-Chia, et al. (2006). Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway. ''Journal of Bacteriology, 188(20)'', 7290-7296. </ref> (in this scene, both Phe residues are magenta). It has also been shown that, after ligand binding, a proton may bind to an acidic residue in the transmembrane domain, which contains an, as yet, putative network of electrostatically interacting residues, the perturbation of which interacting residues leads to a series of conformational changes that result in drug expulsion. Some residues involved in this chain of events are <scene name='Sandbox_100/Asp_407_408_efflux/1'>Asp 407, Asp 408</scene>, <scene name='Sandbox_100/Lys_940_efflux/1'>Lys 940</scene> and <scene name='Sandbox_100/Thr_178_efflux/1'>Thr 978</scene> (red, purple, green, respectively). The precise mechanism of the action of the AcrB efflux subunit (and of the tripartite AcrAB-TolC in general) is still under scrutiny. |
{{Clear}} | {{Clear}} | ||
== Conclusion == | == Conclusion == | ||
| - | As indicated in the explanation of the interaction between Ciprofloxacin and DNA [[Gyrase]], the precise mechanisms of all Ciprofloxacin interactions and transport systems have not been | + | As indicated in the explanation of the interaction between Ciprofloxacin and DNA [[Gyrase]], the precise mechanisms of all Ciprofloxacin interactions and transport systems have not been elaborated completely. However, relevant research, particularly for insight on the precise mechanism for AcrB drug efflux, is underway. Regardless, it is clear that the action of Ciprofloxacin ''in vivo'' is important for the treatment of bacterial infections. The action of Ciprofloxacin on protein function seems to provide a specific field of study that could lead to insight on mechanisms for protein function in general. Thus, Ciprofloxacin is indeed a compound of interest in anticipation of a greater understanding of molecular functions. |
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 15:53, 26 June 2013
| |||||||||||
References
- ↑ CIPRO® (ciprofloxacin hydrochloride) TABLETS - CIPRO® (ciprofloxacin*) ORAL SUSPENSION - Drug Information Packet. Bayer HealthCare Pharmaceuticals. Schering Plough Corporation.
- ↑ CIPRO® (ciprofloxacin hydrochloride) TABLETS - CIPRO® (ciprofloxacin*) ORAL SUSPENSION - Drug Information Packet. Bayer HealthCare Pharmaceuticals. Schering Plough Corporation.
- ↑ 2011. Ciprofloxacin. Medicine Plus. American Society of Health-System Pharmacists Inc. 2011. http://www.nlm.nih.gov/medlineplus/druginfo/meds/a688016.html.
- ↑ 2001. Information on Cipro (Ciprofloxacin Hydrochloride) for Inhalation Anthrax for Consumers: Questions and Answers. Fda.gov. http://www.fda.gov/Drugs/EmergencyPreparedness/BioterrorismandDrugPreparedness/ucm130711.htm. Last updated, 2009.
- ↑ 2011. Ciprofloxacin. Medicine Plus. American Society of Health-System Pharmacists Inc. 2011. http://www.nlm.nih.gov/medlineplus/druginfo/meds/a688016.html.
- ↑ Ciprofloxacin. (2010). Pcm.me. http://pcm.me/ciprofloxacin/.
- ↑ Ciprofloxacin - Activity, Business Aspects/Bayer Pharmaceutical. Encyclopedia.jrank.org. http://encyclopedia.jrank.org/articles/pages/1398940/Ciprofloxacin.html
- ↑ Siegmund, K., et al. (2005). Molecular details of quinolone-DNA interactions: solution structure of an unusually stable DNA duplex with covalently linked nalidixic acid residues and non-covalent complexes derived from it. Nucleic Acids [Research], 33(15), 4838-4848.
- ↑ Peterson, L. (2001). Quinolone-Molecular Structure-Activity Relationships: What We Have Learned About Improving Antimicrobial Activity. Clinical Infectious Diseases, 33(3), S180-S186.
- ↑ Image from: http://cid.oxfordjournals.org/content/33/Supplement_3/S180.full.
- ↑ CIPRO® (ciprofloxacin hydrochloride) TABLETS - CIPRO® (ciprofloxacin*) ORAL SUSPENSION - Drug Information Packet. Bayer HealthCare Pharmaceuticals. Schering Plough Corporation.
- ↑ Molecular weight from Chemexper.com.
- ↑ CIPRO® (ciprofloxacin hydrochloride) TABLETS - CIPRO® (ciprofloxacin*) ORAL SUSPENSION - Drug Information Packet. Bayer HealthCare Pharmaceuticals. Schering Plough Corporation.
- ↑ Image from: http://www.google.com/imgres?imgurl=http://textbookofbacteriology.net/themicrobialworld/cipro.gif&imgrefurl=http://textbookofbacteriology.net/themicrobialworld/control.html&usg=__wtzKLHB3NssfnODEB224br5-Bcw=&h=200&w=250&sz=2&hl=en&start=0&zoom=1&tbnid=o7VT7s6FFIUrWM:&tbnh=160&tbnw=199&ei=Hk10TaypBcL58AbyvIjKDw&prev=/images%3Fq%3Dciprofloxacin%26um%3D1%26hl%3Den%26client%3Dfirefox-a%26sa%3DN%26rls%3Dorg.mozilla:en-US:official%26biw%3D1280%26bih%3D647%26tbs%3Disch:1&um=1&itbs=1&iact=hc&vpx=527&vpy=300&dur=1709&hovh=160&hovw=200&tx=155&ty=82&oei=EU10TcvOCMbdtge5msiLDw&page=1&ndsp=16&ved=1t:429,r:7,s:0.
- ↑ Molecular weight from: CIPRO® (ciprofloxacin hydrochloride) TABLETS - CIPRO® (ciprofloxacin*) ORAL SUSPENSION - Drug Information Packet. Bayer HealthCare Pharmaceuticals. Schering Plough Corporation.
- ↑ Image from: http://www.google.com/imgres?imgurl=http://images.rxlist.com/images/rxlist/ciloxan_s.gif&imgrefurl=http://www.rxlist.com/ciloxan_ophthalmic_ointment-drug.htm&usg=__UqTKseSe8hD85c5RLGIz2_dbAg0=&h=142&w=232&sz=2&hl=en&start=16&zoom=1&tbnid=70Q2WG5hppsQ5M:&tbnh=100&tbnw=164&ei=T010TenMFYH_8Aa6gvDKDw&prev=/images%3Fq%3Dciprofloxacin%26um%3D1%26hl%3Den%26client%3Dfirefox-a%26sa%3DN%26rls%3Dorg.mozilla:en-US:official%26biw%3D1280%26bih%3D647%26tbs%3Disch:10%2C624&um=1&itbs=1&iact=hc&vpx=1064&vpy=399&dur=309&hovh=106&hovw=174&tx=98&ty=76&oei=EU10TcvOCMbdtge5msiLDw&page=2&ndsp=18&ved=1t:429,r:17,s:16&biw=1280&bih=647.
- ↑ Ciprofloxacin. (2010). Pcm.me. http://pcm.me/ciprofloxacin/.
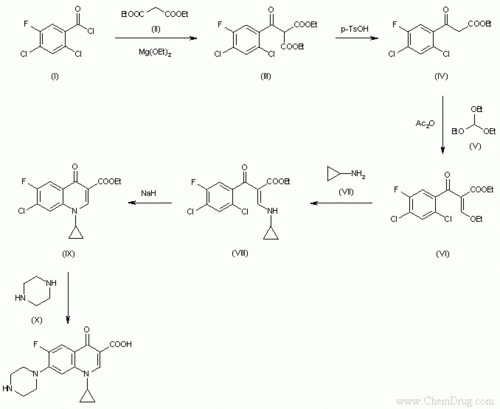
- ↑ Image from: http://www.google.com/imgres?imgurl=http://www.chemdrug.com/databases/SYNTHESIS/SYN/09/09000601a.gif&imgrefurl=http://www.chemdrug.com/databases/8_0_dvpytumicutbciwa.html&usg=__TxiDuzCve6C_crxmcPYTpfW5d4s=&h=555&w=678&sz=6&hl=en&start=0&zoom=1&tbnid=xhquLksJBbMnjM:&tbnh=165&tbnw=201&ei=0Y93TdbGI-yI0QGspa25Bw&prev=/images%3Fq%3Dsynthesis%2Bof%2Bciprofloxacin%26um%3D1%26hl%3Den%26client%3Dfirefox-a%26sa%3DN%26rls%3Dorg.mozilla:en-US:official%26biw%3D1280%26bih%3D647%26tbs%3Disch:1&um=1&itbs=1&iact=hc&vpx=346&vpy=105&dur=63&hovh=203&hovw=248&tx=170&ty=128&oei=0Y93TdbGI-yI0QGspa25Bw&page=1&ndsp=16&ved=1t:429,r:1,s:0
- ↑ Ciprofloxacin Oral - Monograph - Ciprofloxacin Hydrochloride. 2009. Medscape.com. http://www.medscape.com/druginfo/monograph cid=med&drugid=7748&drugname=Ciprofloxacin+Oral&monotype=monograph&secid=8.
- ↑ Su, Chih-Chia, et al. (2006). Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway. Journal of Bacteriology, 188(20), 7290-7296.
- ↑ Husain, F., Nikaido, H. (2010). Substrate path in the AcrB multidrug efflux pump of Escherichia coli. Molecular Microbiology, 78(2), 320-330.
- ↑ Su, Chih-Chia, et al. (2006). Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway. Journal of Bacteriology, 188(20), 7290-7296.
Proteopedia Page Contributors and Editors (what is this?)
John Ripollone, John E. Ripollone, Alexander Berchansky, David Canner