Sandbox Reserved 767
From Proteopedia
| Line 18: | Line 18: | ||
'''Protein Fold and organization of Domain''' | '''Protein Fold and organization of Domain''' | ||
| - | The four identical subunits of TDH are organized into two domains with different sizes. The secondary structure of TDH can be seen in the Figure 3 below. [[Image:QS]. The N-terminal domain is the catalytic domain as it possesses the cofactor of the enzyme, pyridoxal phosphate. This domain consists of residues 1–320 and contains two folding units, N1 and N2. As can be seen in Figure 3, both N1 and N2 subdomains comprise of a collections of four-stranded parallel β sheet with flanking helices. While the N2 subdomain consists of a single contiguous zone of sequence (residues 63–153), N1 is composed of an N2-like core (residues 160–313) with an additional pair of antiparallel strands (residues 34–51) extending the β sheet to six strands. The catalytic domain is homologous (40 identities out of about 280 aligned residues [25]) to the β subunit of tryptophan synthase from Salmonella typhimurium, and the relation of the subdomains is largely as described for that enzyme | + | The four identical subunits of TDH are organized into two domains with different sizes. The secondary structure of TDH can be seen in the Figure 3 below. [[Image:QS]]. The N-terminal domain is the catalytic domain as it possesses the cofactor of the enzyme, pyridoxal phosphate. This domain consists of residues 1–320 and contains two folding units, N1 and N2. As can be seen in Figure 3, both N1 and N2 subdomains comprise of a collections of four-stranded parallel β sheet with flanking helices. While the N2 subdomain consists of a single contiguous zone of sequence (residues 63–153), N1 is composed of an N2-like core (residues 160–313) with an additional pair of antiparallel strands (residues 34–51) extending the β sheet to six strands. The catalytic domain is homologous (40 identities out of about 280 aligned residues [25]) to the β subunit of tryptophan synthase from Salmonella typhimurium, and the relation of the subdomains is largely as described for that enzyme |
'''Cofactor''' | '''Cofactor''' | ||
Revision as of 20:04, 6 December 2013
|
| This Sandbox is Reserved from Sep 25, 2013, through Mar 31, 2014 for use in the course "BCH455/555 Proteins and Molecular Mechanisms" taught by Michael B. Goshe at the North Carolina State University. This reservation includes Sandbox Reserved 299, Sandbox Reserved 300 and Sandbox Reserved 760 through Sandbox Reserved 779. |
To get started:
More help: Help:Editing |
Threonine Dehydratase
Introduction
Threonine dehydratase (TDH) also known as Threonine Ammonia-Lyase and Threonine Deaminase is a tetrameric enzyme of about 200kD that catalyzes the dehydration of threonine to α-ketobutyrate. TDH is the first enzyme in the isoleucine biosynthetic pathway. TDH belongs to the enzyme class EC-4. This enzyme class cleaves C-C, C-O, C-N through hydrolysis or oxidation. TDH [[[EC 4.3.1.19]]] specifically cleaves C-N bonds. TDH was first discovered in E.coli around 1965 but has since been purified from other bacterial sources such as Salmonella typhimurium. TDH possesses 3 domains, one of its cofactor binds to and two regulatory domains.
Structure
Threonine deaminase, purified from Salmonella typhimurium has a molecular weight of about 200kD which exists as tetramer in its native state and consists of four identical subunits. In the absence of the cofactor pyridoxal phosphate, TDH dissociates from its tetrameric form to about 2 individual 100 kD dimers. It is 514 residues in length with a secondary structure that is 37% helical and 17% beta sheet.
Protein Fold and organization of Domain The four identical subunits of TDH are organized into two domains with different sizes. The secondary structure of TDH can be seen in the Figure 3 below. Image:QS. The N-terminal domain is the catalytic domain as it possesses the cofactor of the enzyme, pyridoxal phosphate. This domain consists of residues 1–320 and contains two folding units, N1 and N2. As can be seen in Figure 3, both N1 and N2 subdomains comprise of a collections of four-stranded parallel β sheet with flanking helices. While the N2 subdomain consists of a single contiguous zone of sequence (residues 63–153), N1 is composed of an N2-like core (residues 160–313) with an additional pair of antiparallel strands (residues 34–51) extending the β sheet to six strands. The catalytic domain is homologous (40 identities out of about 280 aligned residues [25]) to the β subunit of tryptophan synthase from Salmonella typhimurium, and the relation of the subdomains is largely as described for that enzyme
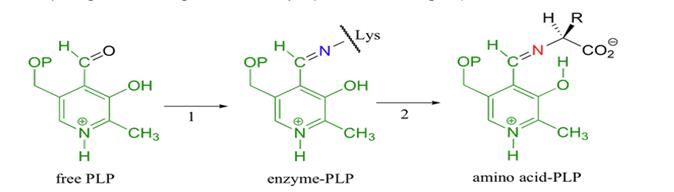
Cofactor For TDH to be catalytically active, pyridoxal phosphate (PLP), its prosthetic group must be attached to TDH. This is initiated when the aldehyde group of PLP forms a Schiff base linkage with the Lys-62 residue of TDH. The electrophilic nitrogen of the pyridine ring of PLP acts as an electron sink where excess electrons can be delocalizes. This effect draws electrons from TDH thereby stabilizing the carbanion intermediate . Below is an illustration of the mechanism of attachment of the cofactor PLP a lysine residue.
Mechanism of Action
Application