This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
3wu2
From Proteopedia
(Difference between revisions)
m (Protected "3wu2" [edit=sysop:move=sysop]) |
|||
| Line 1: | Line 1: | ||
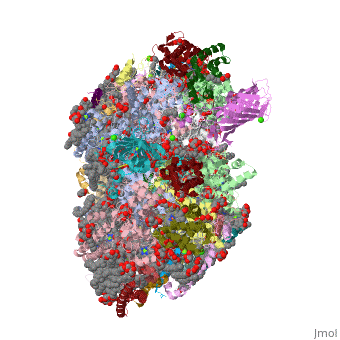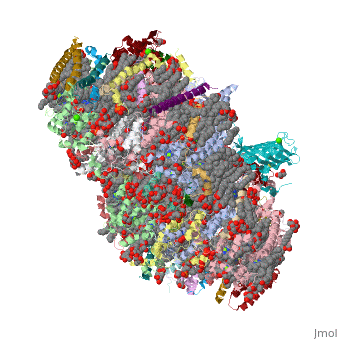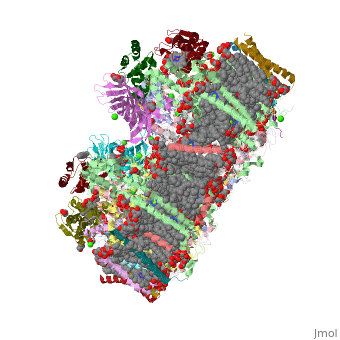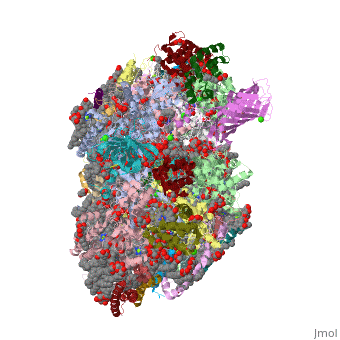
| - | ''' | + | ==Crystal structure analysis of Photosystem II complex== |
| + | <StructureSection load='3wu2' size='340' side='right' caption='[[3wu2]], [[Resolution|resolution]] 1.90Å' scene=''> | ||
| + | == Structural highlights == | ||
| + | <table><tr><td colspan='2'>[[3wu2]] is a 38 chain structure with sequence from [http://en.wikipedia.org/wiki/Thermosynechococcus_vulcanus Thermosynechococcus vulcanus]. This structure supersedes the now removed PDB entry [http://oca.weizmann.ac.il/oca-bin/send-pdb?obs=1&id=3arc 3arc]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3WU2 OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3WU2 FirstGlance]. <br> | ||
| + | </td></tr><tr><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=BCR:BETA-CAROTENE'>BCR</scene>, <scene name='pdbligand=BCT:BICARBONATE+ION'>BCT</scene>, <scene name='pdbligand=CA:CALCIUM+ION'>CA</scene>, <scene name='pdbligand=CL:CHLORIDE+ION'>CL</scene>, <scene name='pdbligand=CLA:CHLOROPHYLL+A'>CLA</scene>, <scene name='pdbligand=DGD:DIGALACTOSYL+DIACYL+GLYCEROL+(DGDG)'>DGD</scene>, <scene name='pdbligand=FE2:FE+(II)+ION'>FE2</scene>, <scene name='pdbligand=GOL:GLYCEROL'>GOL</scene>, <scene name='pdbligand=HEM:PROTOPORPHYRIN+IX+CONTAINING+FE'>HEM</scene>, <scene name='pdbligand=HTG:HEPTYL+1-THIOHEXOPYRANOSIDE'>HTG</scene>, <scene name='pdbligand=LHG:1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE'>LHG</scene>, <scene name='pdbligand=LMG:1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE'>LMG</scene>, <scene name='pdbligand=LMT:DODECYL-BETA-D-MALTOSIDE'>LMT</scene>, <scene name='pdbligand=MG:MAGNESIUM+ION'>MG</scene>, <scene name='pdbligand=OEX:CA-MN4-O5+CLUSTER'>OEX</scene>, <scene name='pdbligand=PHO:PHEOPHYTIN+A'>PHO</scene>, <scene name='pdbligand=PL9:2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE'>PL9</scene>, <scene name='pdbligand=RRX:(3R)-BETA,BETA-CAROTEN-3-OL'>RRX</scene>, <scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene>, <scene name='pdbligand=SQD:1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL'>SQD</scene>, <scene name='pdbligand=UNL:UNKNOWN+LIGAND'>UNL</scene><br> | ||
| + | <tr><td class="sblockLbl"><b>[[Non-Standard_Residue|NonStd Res:]]</b></td><td class="sblockDat"><scene name='pdbligand=FME:N-FORMYLMETHIONINE'>FME</scene>, <scene name='pdbligand=HSK:3-HYDROXY-L-HISTIDINE'>HSK</scene></td></tr> | ||
| + | <tr><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3wu2 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3wu2 OCA], [http://www.rcsb.org/pdb/explore.do?structureId=3wu2 RCSB], [http://www.ebi.ac.uk/pdbsum/3wu2 PDBsum]</span></td></tr> | ||
| + | <table> | ||
| + | <div style="background-color:#fffaf0;"> | ||
| + | == Publication Abstract from PubMed == | ||
| + | Photosystem II is the site of photosynthetic water oxidation and contains 20 subunits with a total molecular mass of 350 kDa. The structure of photosystem II has been reported at resolutions from 3.8 to 2.9 A. These resolutions have provided much information on the arrangement of protein subunits and cofactors but are insufficient to reveal the detailed structure of the catalytic centre of water splitting. Here we report the crystal structure of photosystem II at a resolution of 1.9 A. From our electron density map, we located all of the metal atoms of the Mn(4)CaO(5) cluster, together with all of their ligands. We found that five oxygen atoms served as oxo bridges linking the five metal atoms, and that four water molecules were bound to the Mn(4)CaO(5) cluster; some of them may therefore serve as substrates for dioxygen formation. We identified more than 1,300 water molecules in each photosystem II monomer. Some of them formed extensive hydrogen-bonding networks that may serve as channels for protons, water or oxygen molecules. The determination of the high-resolution structure of photosystem II will allow us to analyse and understand its functions in great detail. | ||
| - | + | Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 A.,Umena Y, Kawakami K, Shen JR, Kamiya N Nature. 2011 May 5;473(7345):55-60. Epub 2011 Apr 17. PMID:21499260<ref>PMID:21499260</ref> | |
| - | + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |
| - | + | </div> | |
| - | + | == References == | |
| + | <references/> | ||
| + | __TOC__ | ||
| + | </StructureSection> | ||
| + | [[Category: Thermosynechococcus vulcanus]] | ||
| + | [[Category: Kamiya, N.]] | ||
| + | [[Category: Kawakami, K.]] | ||
| + | [[Category: Shen, J R.]] | ||
| + | [[Category: Umena, Y.]] | ||
| + | [[Category: Calcium binding]] | ||
| + | [[Category: Chloride binding]] | ||
| + | [[Category: Electron transport]] | ||
| + | [[Category: Formylation]] | ||
| + | [[Category: Hydroxylation]] | ||
| + | [[Category: Iron binding]] | ||
| + | [[Category: Manganese binding]] | ||
| + | [[Category: Membrane complex]] | ||
| + | [[Category: Oxygen evolving]] | ||
| + | [[Category: Photosynthesis]] | ||
| + | [[Category: Photosystem]] | ||
| + | [[Category: Psii]] | ||
| + | [[Category: Thylakoid membrane]] | ||
| + | [[Category: Transmembrane alpha-helix]] | ||
| + | [[Category: Water splitting]] | ||
Revision as of 05:43, 4 September 2014
Crystal structure analysis of Photosystem II complex
| |||||||||||
Categories: Thermosynechococcus vulcanus | Kamiya, N. | Kawakami, K. | Shen, J R. | Umena, Y. | Calcium binding | Chloride binding | Electron transport | Formylation | Hydroxylation | Iron binding | Manganese binding | Membrane complex | Oxygen evolving | Photosynthesis | Photosystem | Psii | Thylakoid membrane | Transmembrane alpha-helix | Water splitting