This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 930
From Proteopedia
| Line 7: | Line 7: | ||
[[Image:Actin myosin anim.gif|400px|left|thumb| Figure 1. The movement of myosin motor domain on actin filament]] | [[Image:Actin myosin anim.gif|400px|left|thumb| Figure 1. The movement of myosin motor domain on actin filament]] | ||
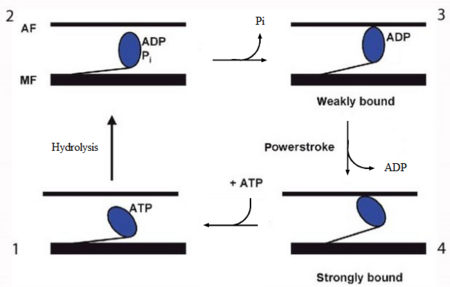
[[Image:myosin.png|450px|right|thumb| Figure 2. The contractile cycle of the myosin head]] | [[Image:myosin.png|450px|right|thumb| Figure 2. The contractile cycle of the myosin head]] | ||
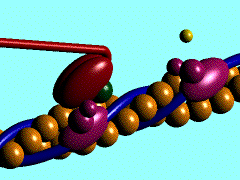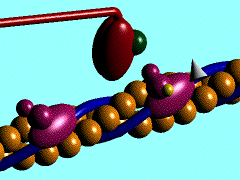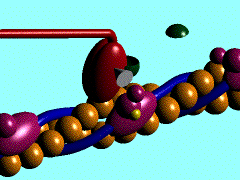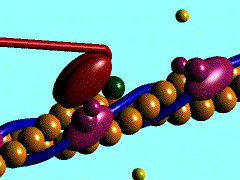
| - | In the striated muscle the actin and myosin proteins form ordered basic units called sarcomeres. Muscle contraction is achieved by the mechanical sliding of myosin filament (thick filament) along the actin filament (thin filament), Fig. 1. The major constituent of the myosin filament is myosin, a motor protein responsible for converting chemical energy to mechanical movement. In the presence of Ca<sup>2+</sup> and Mg<sup>2+</sup>, myosin is able to cyclically bind ATP and hydrolyse it to ADP + P<sub>i</sub> , triggering subsequent myosin-actin detachment, reattachment and power stroke, so called contractile reaction (Fig.2). | + | In the striated muscle the actin and myosin proteins form ordered basic units called sarcomeres. Muscle contraction is achieved by the mechanical sliding of myosin filament (thick filament) along the actin filament (thin filament), Fig. 1. The major constituent of the myosin filament is myosin, a motor protein responsible for converting chemical energy to mechanical movement. In the presence of Ca<sup>2+</sup> and Mg<sup>2+</sup>, myosin is able to cyclically bind ATP and hydrolyse it to ADP + P<sub>i</sub> , thus triggering subsequent myosin-actin detachment, reattachment and power stroke, the so called contractile reaction (Fig.2). |
| Line 23: | Line 23: | ||
==Introduction of the Myosin head S1 == | ==Introduction of the Myosin head S1 == | ||
<StructureSection load='1B7T' size='450' frame='true' side='right' caption='Myosin subfragment 1' scene='57/579700/Whole_structure/3'> | <StructureSection load='1B7T' size='450' frame='true' side='right' caption='Myosin subfragment 1' scene='57/579700/Whole_structure/3'> | ||
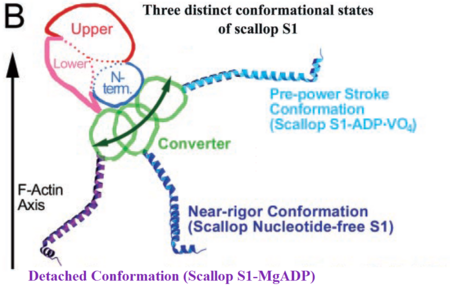
| - | Myosin is a large asymmetric molecule with a MW of about 500,000 kDa. It consist of two globular head domains termed myosin subfragment 1 (S1), one neck subfragment 2 (S2) and a light meromyosin tail (LMM) <ref>PMID: 8203020</ref>. Myosin S1 unit comprises of a motor domain (MD) and a lever arm (Fig.3) | + | Myosin is a large asymmetric molecule with a MW of about 500,000 kDa. It consist of two globular head domains termed myosin subfragment 1 (S1), one neck subfragment 2 (S2) and a light meromyosin tail (LMM) <ref>PMID: 8203020</ref>. Myosin S1 unit comprises of a motor domain (MD) and a lever arm (Fig.3) By 2000 scallop myosin S1 has been determined in three different conformations of the contractile cycle, corresponding to the following structures <ref>PMID: 11016966</ref><ref>PMID: 10338210</ref>: |
| - | • S1 nucleotide-free state corresponding to the rigor | + | • S1 nucleotide-free state corresponding to the near-rigor conformation of myosin [http://www.pdb.org/pdb/explore/explore.do?structureId=1DFK 1DFK] |
| - | • S1 Mg-ADP.VO4 state corresponding to the pre-power stroke | + | • S1 Mg-ADP.VO4 state corresponding to the pre-power stroke conformation [http://www.pdb.org/pdb/explore/explore.do?structureId=1kk8 1KK8] |
• S1 Mg-ADP state corresponding to the myosin detached state [http://www.rcsb.org/pdb/explore/explore.do?structureId=1b7t 1B7T] | • S1 Mg-ADP state corresponding to the myosin detached state [http://www.rcsb.org/pdb/explore/explore.do?structureId=1b7t 1B7T] | ||
| - | + | Comparing the available crystal structures of different myosin the S1 unit enable us to understand the conformational changes within the motor domain during the contractile cycle. Here mainly the structure and function of the MD in the S1 Mg-ADP.VO4 (pre power stroke) state will be discussed. | |
| + | |||
| + | depending on the nucleotide content in the active site. Here mainly the structure and function of MD relevant in the S1 Mg-ADP (pre power stroke) state will be discussed. | ||
==The subdomains of the motor domain== | ==The subdomains of the motor domain== | ||
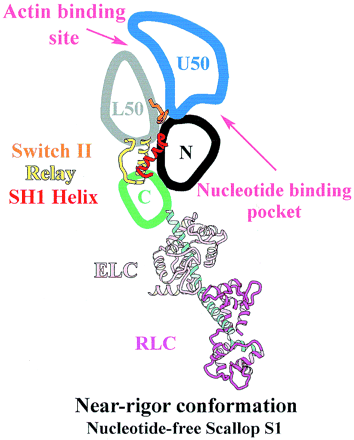
[[Image:SUBDOMAINS.gif|400px|right|thumb|Figure 3. The subdomains on myosin S1 unit]] | [[Image:SUBDOMAINS.gif|400px|right|thumb|Figure 3. The subdomains on myosin S1 unit]] | ||
| - | The MD of S1 unit is most frequently described as consisting of | + | The MD of the scallop S1 unit is most frequently described as consisting of four subdomains: the converter, the N-terminal subdomain, and the upper and lower 50-kDa subdomains <ref>PMID: 10338210</ref>. They are linked together by three single-stranded joints termed the switch II (residues IIe-461 to Asn-470), the relay (residues Asn-489 to Asp-519), and SH1 helix (residues Cys-693 to Phe-707)(Fig. 3) <ref>PMID: 11016966</ref>. |
| - | • Of the MD subdomains the converter | + | • Of the MD subdomains the converter changes its position the most during the contractile cycle. Connection of the converter and the lever arm allows relatively small changes in the converter to be greatly amplified in the lever arm (Fig. 4) |
• Upper 50-kDa subdomain and N-terminal subdomain form the nucleotide binding pocket | • Upper 50-kDa subdomain and N-terminal subdomain form the nucleotide binding pocket | ||
| Line 44: | Line 46: | ||
• Lower and upper 50-kDa subdomains form the interface where actin can bind <ref>PMID: 11016966</ref> | • Lower and upper 50-kDa subdomains form the interface where actin can bind <ref>PMID: 11016966</ref> | ||
| - | Conformational changes in the flexible joints coordinate rearrangements of | + | Conformational changes in the flexible joints coordinate rearrangements of the four MD subdomains enabling the transition between different myosin S1 conformations in the actomyosin contractile cycle, during which S1 traduces ATP hydrolysis to mechanical work. The different conformational states of myosin are termed strong or weak actin-binding states <ref>PMID: 15184651</ref>. |
| Line 51: | Line 53: | ||
==Nucleotide binding pocket: ADP + Mg<sup>2+</sup>== | ==Nucleotide binding pocket: ADP + Mg<sup>2+</sup>== | ||
| - | The nucleotide-binding pocket is located at the interface of the 50 kDa upper subdomain and the N-terminal subdomain, which is opposite to a deep cleft that bisects the actin-binding domain ( | + | The nucleotide-binding pocket is located at the interface of the 50 kDa upper subdomain and the N-terminal subdomain <ref>PMID: 15184651</ref>, which is opposite to a deep cleft that bisects the actin-binding domain (Fig. 3).This part of protein involves an arrangement of a secondary structure mainly around the parallel 7-stranded <scene name='57/579700/Strands/1'> β-sheet</scene> (reference 1). Loops extending from b-strands interact with the adenine nucleotide. |
| - | <scene name='57/579700/Adp/5'>ADP</scene> forms hydrogen bonds with the amino acid side | + | <scene name='57/579700/Adp/5'>ADP</scene> forms hydrogen bonds with the amino acid side chains around it, <scene name='57/579700/Mg/5'>Mg2+</scene> coordinates with residues Thr183, Ser 241 of the heavy chain, O1B and O3B from ADP and three water molecules (Scene) as well. The hydrogen bonds between ADP and the amino acid residues together with the Mg2+ keep ADP in the nucleotide-binding pocket. |
| - | In the contractile cycle | + | In the contractile cycle ATP binding causes a conformational change, which detaches the myosin S1 unit from actin. Then the active site closes, and ATP is hydrolysed to Pi and ADP, leading to the subsequent reattachment of the S1 with the actin. The conformational changes of the acting-binding pocket and the opening and closing of the nucleotide-binding pocket cause the strong and weak acting binding states of myosin, allowing muscle contraction. |
| Line 67: | Line 69: | ||
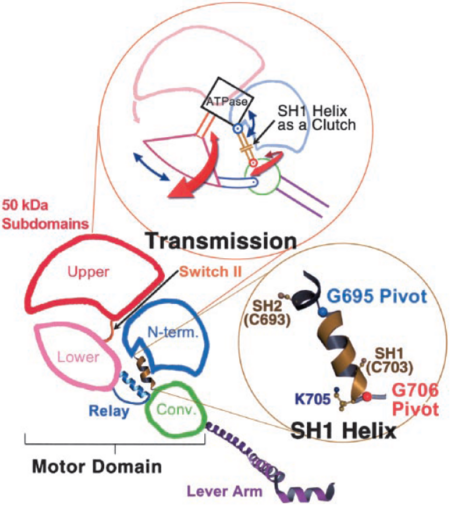
The 50-kDa upper and lower subdomains as well as the converter control the motor function of the myosin head by rotating around the N-terminal subdomain (Fig 5). The rotations depend on the conformational changes of the 3 joints; switch II, SH1 helix region, and relay. <ref>PMID: 15184651</ref> The joints work together in the transition between the different conformational states of MD to control the overall organization of the myosin head. They also allow communication between the nucleotide-bonding pocket, acting-binding interface and the lever arm <ref>PMID: 11016966</ref>. | The 50-kDa upper and lower subdomains as well as the converter control the motor function of the myosin head by rotating around the N-terminal subdomain (Fig 5). The rotations depend on the conformational changes of the 3 joints; switch II, SH1 helix region, and relay. <ref>PMID: 15184651</ref> The joints work together in the transition between the different conformational states of MD to control the overall organization of the myosin head. They also allow communication between the nucleotide-bonding pocket, acting-binding interface and the lever arm <ref>PMID: 11016966</ref>. | ||
| - | Switch II, a catalytic loop of the nucleotide-binding pocket, moves in and out of the nucleotide-binding pocket during enzymatic activity. It is responsible for the unwinding of SH1 helix, along with the conformational changes caused by nucleotide binding. | + | Switch II, a catalytic loop of the nucleotide-binding pocket, moves in and out of the nucleotide-binding pocket during enzymatic activity. It is responsible for the unwinding of SH1 helix, along with the conformational changes caused by nucleotide binding. As SH1 unwinds, by rotating around two pivots (G695, G706, Fig. 5), it uncouples the converter/lever module from the MD <ref>PMID: 12297624</ref>. Movement of the converter is controlled by the relay joint. The converter/relay module attains different conformations changing the position of the lever arm (Fig. 4) and thus giving rise to the different states in the actomyosin cycle <ref>PMID: 11016966</ref>. |
Taking part in the organization of the different conformations of the contractile cycle is also the so called switch I, which is a second catalytic loop of the nucleotide-binding pocket. <ref>PMID: 11016966</ref> Switch II forms a specific salt bridge and hydrogen bond interactions with switch I that stabilize the pre-power stroke state <ref>PMID: 12297624</ref>. | Taking part in the organization of the different conformations of the contractile cycle is also the so called switch I, which is a second catalytic loop of the nucleotide-binding pocket. <ref>PMID: 11016966</ref> Switch II forms a specific salt bridge and hydrogen bond interactions with switch I that stabilize the pre-power stroke state <ref>PMID: 12297624</ref>. | ||
| - | In the pre-power stroke conformation of the MD, switch II interacts with the nucleotide-binding pocket and forms the stabilizing hydrogen bond interactions and a salt bridge with switch I. <ref>PMID: 12297624</ref><ref>PMID: 15184651</ref> Rotation of the 50-kDa upper subdomain away from the N-terminal subdomain pulls switches I and II apart breaking the protein- nucleotide interactions between switch I and ADP, as well as changing the conformation of switch II. These changes result in closing of the actin-binding site and opening of the nucleotide-binding pocket, leading to MgADP release. At the same | + | In the pre-power stroke conformation of the MD, switch II interacts with the nucleotide-binding pocket and forms the stabilizing hydrogen bond interactions and a salt bridge with switch I. <ref>PMID: 12297624</ref><ref>PMID: 15184651</ref> Rotation of the 50-kDa upper subdomain away from the N-terminal subdomain pulls switches I and II apart breaking the protein- nucleotide interactions between switch I and ADP, as well as changing the conformation of switch II. These changes result in closing of the actin-binding site and opening of the nucleotide-binding pocket, leading to MgADP release. At the same time SH1 helix is unwound and the lever arm is able to change its position enabling sliding of myosin through the actin filament <ref>PMID: 12297624</ref>. |
Revision as of 11:02, 17 May 2014
| This Sandbox is Reserved from 01/04/2014, through 30/06/2014 for use in the course "510042. Protein structure, function and folding" taught by Prof Adrian Goldman, Tommi Kajander, Taru Meri, Konstantin Kogan and Juho Kellosalo at the University of Helsinki. This reservation includes Sandbox Reserved 923 through Sandbox Reserved 947. |
To get started:
More help: Help:Editing |
Contents |
Scallop myosin head in its pre power stroke state
Introduction
In the striated muscle the actin and myosin proteins form ordered basic units called sarcomeres. Muscle contraction is achieved by the mechanical sliding of myosin filament (thick filament) along the actin filament (thin filament), Fig. 1. The major constituent of the myosin filament is myosin, a motor protein responsible for converting chemical energy to mechanical movement. In the presence of Ca2+ and Mg2+, myosin is able to cyclically bind ATP and hydrolyse it to ADP + Pi , thus triggering subsequent myosin-actin detachment, reattachment and power stroke, the so called contractile reaction (Fig.2).
.
Introduction of the Myosin head S1
| |||||||||||
References
- ↑ Rayment I, Holden HM. The three-dimensional structure of a molecular motor. Trends Biochem Sci. 1994 Mar;19(3):129-34. PMID:8203020
- ↑ Houdusse A, Szent-Gyorgyi AG, Cohen C. Three conformational states of scallop myosin S1. Proc Natl Acad Sci U S A. 2000 Oct 10;97(21):11238-43. PMID:11016966 doi:10.1073/pnas.200376897
- ↑ Houdusse A, Kalabokis VN, Himmel D, Szent-Gyorgyi AG, Cohen C. Atomic structure of scallop myosin subfragment S1 complexed with MgADP: a novel conformation of the myosin head. Cell. 1999 May 14;97(4):459-70. PMID:10338210
- ↑ Houdusse A, Kalabokis VN, Himmel D, Szent-Gyorgyi AG, Cohen C. Atomic structure of scallop myosin subfragment S1 complexed with MgADP: a novel conformation of the myosin head. Cell. 1999 May 14;97(4):459-70. PMID:10338210
- ↑ Houdusse A, Szent-Gyorgyi AG, Cohen C. Three conformational states of scallop myosin S1. Proc Natl Acad Sci U S A. 2000 Oct 10;97(21):11238-43. PMID:11016966 doi:10.1073/pnas.200376897
- ↑ Houdusse A, Szent-Gyorgyi AG, Cohen C. Three conformational states of scallop myosin S1. Proc Natl Acad Sci U S A. 2000 Oct 10;97(21):11238-43. PMID:11016966 doi:10.1073/pnas.200376897
- ↑ Risal D, Gourinath S, Himmel DM, Szent-Gyorgyi AG, Cohen C. Myosin subfragment 1 structures reveal a partially bound nucleotide and a complex salt bridge that helps couple nucleotide and actin binding. Proc Natl Acad Sci U S A. 2004 Jun 15;101(24):8930-5. Epub 2004 Jun 7. PMID:15184651 doi:10.1073/pnas.0403002101
- ↑ Risal D, Gourinath S, Himmel DM, Szent-Gyorgyi AG, Cohen C. Myosin subfragment 1 structures reveal a partially bound nucleotide and a complex salt bridge that helps couple nucleotide and actin binding. Proc Natl Acad Sci U S A. 2004 Jun 15;101(24):8930-5. Epub 2004 Jun 7. PMID:15184651 doi:10.1073/pnas.0403002101
- ↑ Houdusse A, Szent-Gyorgyi AG, Cohen C. Three conformational states of scallop myosin S1. Proc Natl Acad Sci U S A. 2000 Oct 10;97(21):11238-43. PMID:11016966 doi:10.1073/pnas.200376897
- ↑ Risal D, Gourinath S, Himmel DM, Szent-Gyorgyi AG, Cohen C. Myosin subfragment 1 structures reveal a partially bound nucleotide and a complex salt bridge that helps couple nucleotide and actin binding. Proc Natl Acad Sci U S A. 2004 Jun 15;101(24):8930-5. Epub 2004 Jun 7. PMID:15184651 doi:10.1073/pnas.0403002101
- ↑ Houdusse A, Szent-Gyorgyi AG, Cohen C. Three conformational states of scallop myosin S1. Proc Natl Acad Sci U S A. 2000 Oct 10;97(21):11238-43. PMID:11016966 doi:10.1073/pnas.200376897
- ↑ Himmel DM, Gourinath S, Reshetnikova L, Shen Y, Szent-Gyorgyi AG, Cohen C. Crystallographic findings on the internally uncoupled and near-rigor states of myosin: further insights into the mechanics of the motor. Proc Natl Acad Sci U S A. 2002 Oct 1;99(20):12645-50. Epub 2002 Sep 24. PMID:12297624 doi:10.1073/pnas.202476799
- ↑ Houdusse A, Szent-Gyorgyi AG, Cohen C. Three conformational states of scallop myosin S1. Proc Natl Acad Sci U S A. 2000 Oct 10;97(21):11238-43. PMID:11016966 doi:10.1073/pnas.200376897
- ↑ Houdusse A, Szent-Gyorgyi AG, Cohen C. Three conformational states of scallop myosin S1. Proc Natl Acad Sci U S A. 2000 Oct 10;97(21):11238-43. PMID:11016966 doi:10.1073/pnas.200376897
- ↑ Himmel DM, Gourinath S, Reshetnikova L, Shen Y, Szent-Gyorgyi AG, Cohen C. Crystallographic findings on the internally uncoupled and near-rigor states of myosin: further insights into the mechanics of the motor. Proc Natl Acad Sci U S A. 2002 Oct 1;99(20):12645-50. Epub 2002 Sep 24. PMID:12297624 doi:10.1073/pnas.202476799
- ↑ Himmel DM, Gourinath S, Reshetnikova L, Shen Y, Szent-Gyorgyi AG, Cohen C. Crystallographic findings on the internally uncoupled and near-rigor states of myosin: further insights into the mechanics of the motor. Proc Natl Acad Sci U S A. 2002 Oct 1;99(20):12645-50. Epub 2002 Sep 24. PMID:12297624 doi:10.1073/pnas.202476799
- ↑ Risal D, Gourinath S, Himmel DM, Szent-Gyorgyi AG, Cohen C. Myosin subfragment 1 structures reveal a partially bound nucleotide and a complex salt bridge that helps couple nucleotide and actin binding. Proc Natl Acad Sci U S A. 2004 Jun 15;101(24):8930-5. Epub 2004 Jun 7. PMID:15184651 doi:10.1073/pnas.0403002101
- ↑ Himmel DM, Gourinath S, Reshetnikova L, Shen Y, Szent-Gyorgyi AG, Cohen C. Crystallographic findings on the internally uncoupled and near-rigor states of myosin: further insights into the mechanics of the motor. Proc Natl Acad Sci U S A. 2002 Oct 1;99(20):12645-50. Epub 2002 Sep 24. PMID:12297624 doi:10.1073/pnas.202476799