We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 967
From Proteopedia
(Difference between revisions)
| Line 12: | Line 12: | ||
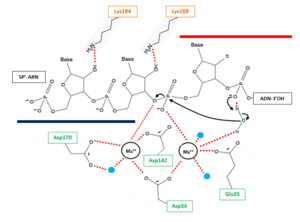
Indeed, ribonucleotides are wrongly incorporated into DNA during DNA replication at a frequency of about 2 ribonucleotides per kb. With such frequency, these errors are by far the most abundant threat of DNA damaging. Hence, a correction is essential to the preservation of DNA integrity: the most common correction mechanism involves RNases H2 and is called Ribonucleotide Excision Repair (RER). The incorporation of ribonucleotides in DNA produce DNA•RNAfew•DNA/DNA hybrids from which the few misincorporated ribonucleotides can be removed by an RNase H2<ref> Sparks, Justin L., Hyongi Chon, Susana M. Cerritelli, Thomas A. Kunkel, Erik Johansson, Robert J. Crouch, and Peter M. Burgers. “RNase H2-Initiated Ribonucleotide Excision Repair.” Molecular Cell 47, no. 6 (September 28, 2012): 980–86. doi:10.1016/j.molcel.2012.06.035.</ref>. | Indeed, ribonucleotides are wrongly incorporated into DNA during DNA replication at a frequency of about 2 ribonucleotides per kb. With such frequency, these errors are by far the most abundant threat of DNA damaging. Hence, a correction is essential to the preservation of DNA integrity: the most common correction mechanism involves RNases H2 and is called Ribonucleotide Excision Repair (RER). The incorporation of ribonucleotides in DNA produce DNA•RNAfew•DNA/DNA hybrids from which the few misincorporated ribonucleotides can be removed by an RNase H2<ref> Sparks, Justin L., Hyongi Chon, Susana M. Cerritelli, Thomas A. Kunkel, Erik Johansson, Robert J. Crouch, and Peter M. Burgers. “RNase H2-Initiated Ribonucleotide Excision Repair.” Molecular Cell 47, no. 6 (September 28, 2012): 980–86. doi:10.1016/j.molcel.2012.06.035.</ref>. | ||
| - | This repair activity is guided by the interaction between C-terminus of RNase H2B protein and the DNA clamp PCNA. This interaction occurs through a hydrophobic conserved peptide motif called the PCNA interaction peptide PIP (PIP-box: Residues 294 to 301 MKSIDTFF of H2B protein) that interacts with a hydrophobic groove near the PCNA C-terminus. This interaction allows RNase H2 to scan DNA for misincorporated ribonucleotides which makes the Ribonucleotide Excision Repair more efficient<ref> Bubeck, Doryen, Martin A. M. Reijns, Stephen C. Graham, Katy R. Astell, E. Yvonne Jones, and Andrew P. Jackson. “PCNA Directs Type 2 RNase H Activity on DNA Replication and Repair Substrates.” Nucleic Acids Research 39, no. 9 (May 2011): 3652–66. | + | This repair activity is guided by the interaction between C-terminus of RNase H2B protein and the DNA clamp PCNA. This interaction occurs through a hydrophobic conserved peptide motif called the PCNA interaction peptide PIP (PIP-box: Residues 294 to 301 MKSIDTFF of H2B protein) that interacts with a hydrophobic groove near the PCNA C-terminus. This interaction allows RNase H2 to scan DNA for misincorporated ribonucleotides which makes the Ribonucleotide Excision Repair more efficient<ref> Bubeck, Doryen, Martin A. M. Reijns, Stephen C. Graham, Katy R. Astell, E. Yvonne Jones, and Andrew P. Jackson. “PCNA Directs Type 2 RNase H Activity on DNA Replication and Repair Substrates.” Nucleic Acids Research 39, no. 9 (May 2011): 3652–66. http://dx.doi.org/10.1093/nar/gkq980 doi:10.1093/nar/gkq980.</ref>. |
Furthermore, ''in vitro'' studies have shown that RNases H2 is likely to be involved in the removal of RNA primer from Okazaki fragment produced during the synthesis of the lagging strand in DNA replication since Okazaki fragment are RNA•DNA/DNA hybrids ('''Figure 1B'''). | Furthermore, ''in vitro'' studies have shown that RNases H2 is likely to be involved in the removal of RNA primer from Okazaki fragment produced during the synthesis of the lagging strand in DNA replication since Okazaki fragment are RNA•DNA/DNA hybrids ('''Figure 1B'''). | ||
Revision as of 16:40, 8 January 2015
| This Sandbox is Reserved from 15/11/2014, through 15/05/2015 for use in the course "Biomolecule" taught by Bruno Kieffer at the Strasbourg University. This reservation includes Sandbox Reserved 951 through Sandbox Reserved 975. |
To get started:
More help: Help:Editing |
Structure of the Mouse RNase H2 Complex
| |||||||||||
References
- ↑ http://genome-euro.ucsc.edu/cgi-bin/hgTracks?clade=mammal&org=Mouse&db=mm10&position=RnaseH2&hgt.positionInput=RnaseH2&hgt.suggestTrack=knownGene&Submit=submit&hgsid=201143152_yP1Xd4bMnHS7DV0d3VcqpDSxzzuQ&pix=1563
- ↑ Rychlik, Monika P., Hyongi Chon, Susana M. Cerritelli, Paulina Klimek, Robert J. Crouch, and Marcin Nowotny. “Crystal Structures of RNase H2 in Complex with Nucleic Acid Reveal the Mechanism of RNA-DNA Junction Recognition and Cleavage.” Molecular Cell 40, no. 4 (November 24, 2010): 658–70. doi:10.1016/j.molcel.2010.11.001.
- ↑ Sparks, Justin L., Hyongi Chon, Susana M. Cerritelli, Thomas A. Kunkel, Erik Johansson, Robert J. Crouch, and Peter M. Burgers. “RNase H2-Initiated Ribonucleotide Excision Repair.” Molecular Cell 47, no. 6 (September 28, 2012): 980–86. doi:10.1016/j.molcel.2012.06.035.
- ↑ Bubeck, Doryen, Martin A. M. Reijns, Stephen C. Graham, Katy R. Astell, E. Yvonne Jones, and Andrew P. Jackson. “PCNA Directs Type 2 RNase H Activity on DNA Replication and Repair Substrates.” Nucleic Acids Research 39, no. 9 (May 2011): 3652–66. http://dx.doi.org/10.1093/nar/gkq980 doi:10.1093/nar/gkq980.