Squamous cell carcinoma antigen 1 is a tumor associated protein of squamous cell carcinoma of various organs. SCCA1 was originally purified from SCC of the uterine cervix [1]. SCCA1 is a tumor marker to detect malignant tumor and to understand biological behaviors of squamous cells. SCCA1 is classified as a serine protease inhibitor called serpin B3. It also inhibits chymotripsin, cathepsin L, K and S and papain like cysteine proteases. In the case of tumor development SCCA 1 inhibits NK cells(natural killer), TNFalfa and apoptosis of tumor cells induced by treatment. It can also play a role in tumor growth. The chromosomal location is the locus 18q21.3.[2]
Structure
General structure
Serpins are a superfamily of proteins wich are functionally distinct but structurally conserved. [3]
SerpinB3 means serin protease inhibitor, clade B (for ovalbumin), member 3. The particularity of Serpin B3 is to target proteases which have a nucleophilic cysteine instead of serine in their catalytic site.
SCCA1 is a [4]. has three β sheets termed , and and [5] [6].
The most important domain of SCCA1 is an exposed region of 20 amino acids near the C terminus named the reactive center loop (). The amino-acids of are very conservated for SCCA1 and allow the specificity interaction between the inhibitor and the target protease[7].
Conformational changes of serpins
The inhibitory members of serpin family undergo an unusual conformational change, the Stressed to Relaxed transition. This structural transition causes the insertion into thereby the forms an extra β strand . The serpin conformational change is essential for the inhibition mechanism of proteases. wich belong to a consensus sequence for inhibitory serpins are thought to permit the insertion of the into the .[8]
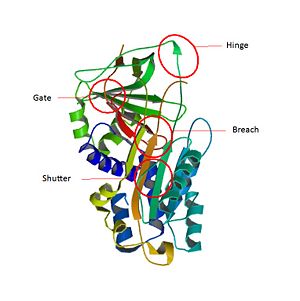
Key regions able to control and modulate the conformational change of RCL Main SCCA1 function
Cysteine proteases mecanism
When a cystein protease attacks a substrate, the peptide bond cleavage is catalyzed. Firstly, the catalytic cysteine performs a nucleophilic attack on the substrate peptide bond. Thus the new N-terminusis is released and a new bond between the enzyme and the substrate is created forming a covalent enzyme-substrate complex called the acyl enzyme intermediate. Finally, this new bond is hydrolysed and the new C-terminus is released.
Protease inhibition
The of SCCA1 acts as a substrate for its target protease.
The is cleaved at a scissile bond between two residues . The are the necessary condition for target protease specificity. Indeed, the mutation of these residues results destroys inhibitory activity. The protease recognize that allow its docking.
[9] [10]
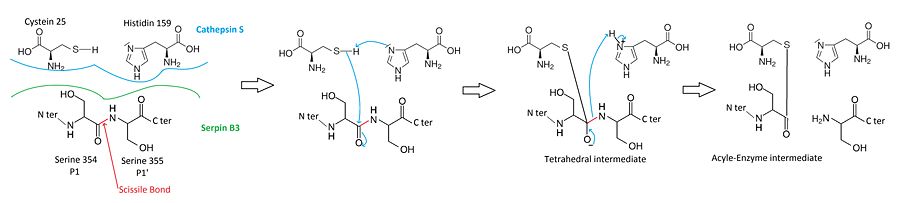
Mecanism of cathepsin S-inhibition by serpin B3
Before the hydrolysis of the acyl-enzyme intermediate, the serpin rapidly undergoes from Stressed to Relaxed transition. The remains covalently attached to the protease, however the protease is moved from the top to the bottom of SCCA1. [11]
The increase of the SCCA1 residing in the cytosol of squamous carcinoma cell may protect the tumor by neutralizing harmful proteases.
The SCCA1 a tumor marker
Cancer is characterized by the abnormal proliferation of a cellular clone that will form a tumor in a tissue. Tumor cells can migrate to the serum or urin and invade other tissues. Cancer is caused by damaged genes. Cancer can have several origins due to exogenous factors (tobacco, alcohol, UV) or endogenous factors (failure in DNA repair).
Tumor markers are compounds present in abnormal concentration in serum or urine in patients who develop a malignant tumor. Nevertheless tumor markers can appear in people who do not suffer from cancer or at low concentration in sick patients, this is called the false negative or false positive. Tumor markers are used to detect, prevent, diagnose, predict, determine, prognostic and therapeutic monitoring. Tumor markers need to be specific and sensitive. The dosage of several markers is necessary to establish the success or the failure of a treatment.
The SCCA is secreted by the tumor itself, it is a marker of mature cells. The SCCA is a glycoprotein present in the epithelium cells and released in the serum during epidermoid cervical cancer but also in epidermoid cancers suchh as lung, mouth, larynx, pharynx and esophagus. The cooncentration threshold is inferior at 1.5 µg for healthy patient.[12]
SCCA role as a tumor marker
SCCA is particularly used for the detecction of cancer of the uterine cervix. The correlation between SCCA concentration and lung tumor was proved. SCCA concentration increases in the presence of epidermoid lung tumor, independently of the differentiation state of the tumor[13]. SCCA is especially used to prognostic and follow the effects of the treatment in the lung cancer as second tumor marker [14]
. High concentration of SCCA in the blood suggests the epithelial cells direct serpin activity to blood. This pathway is an active secretory process[15].
SCCA and cancer
SCCA is not specific of one type of cancer. It can be associated to mild broncho-pulmonarypathology, mild skin pathology. It does not depend on Tobacco consumption. SCCA is associated to cancer and non-malignant kidney pathology. It is quantified by immuno-analyzes, its half-life is 3 days.
-In cervix cancer :
The SCCA increase is linked to the tumor weight and state of disease. Nevertheless 40 % of patients suffering from cervix cancer have a high SCCA blood concentration, it is not use for screening. An increase of the initial rate can be a sign of disease recurrence or persistence. It allows to follow the treatment efficiency such as chemotherapy, radiotherapy in patients.
-In epidermoid bronchopulmonary cancer :
SCCA is not used for screening. [16]
Interaction
Hepatite B virus (HBV) interaction
The SCCA may play a role of cellular receptor for hepatitis B virus. The SCCA expression enhances the binding and internalization of hepatitis B virus with hepatocyte or non-hepatocytes origin cells. The transfection of SCCA in hepatocyte generates more viruses DNA in infected cells. Besides the virus bound to transfected cell is protected against degradation by trypsin thanks to a partial internalization. The binding between HBV and hepatocytes is more marked than for the others types of cells like COS-7 (kidney cells of monkey transformed by antibody T of SV40). The binding complex of cells COS-7 with HBV seems to be more complex. The low density lipoprotein receptor-related protein (LRP) mediates the clearance of serpin-enzyme complex, the LRP may not enhance virus binding to transfected cells. SCCA may be a co-receptor for HBV, virus binding to the transfected cells doesn’t depend on the proteinase inhibitor function or the interaction receptor LRP but it may depend on of SCCA. [17]
JNK1 interaction
SCCA1 acts as an inhibitor of UV-induced apoptosis by suppressing the activity of JNK1 (c-Jun NH2-terminal kinase). It is known that JNK1 is responsible for UV-induced apoptotic cell death and SCCA-1 is up-regulated in UV-irradiated and sun-exposed cells. SCCA1 binds to phosphorylated JNK1 and is transferred into the nucleus after UV irradiation.
[18]
Indeed the () of SCCA1 is an external and flexible domain of SCCA1. When the is truncated, the inhibitory effect of SCCA1 on the kinase activity of JNK1 is lost.
Moreover, a mutant protein created by replacing in even retains the suppressive activity against JNK1 but the inhibitory proteinase activity is reduced. This indicates that exposed is essential for the JNK1 inhibitory activity and that JNK1 interaction site is different from the protease site. Actually, JNK1 may binds the region, excepting the protease binding site, or the center of . [19]
Disease
Asthma is characterized by an obstruction of the interior respiratory tract and an excessive mucus secretion.[20] Experiments were performed on mice, mice lacking SerpinB3 showed a decrease of the mucus secretion. As a result serpinB3 may have a role in mucus hypersecretion in a house dust mist model of asthma. The SPDEF ( SAM pointed domain containing ETS transcription factor) expression causes the hyperplasia of goblet cell. The hyperplasia designates the abnormal augmentation of cells number in a tissue, the subexpression of goblet cells m ay induce cancer. Serpin B3 increase SPDEF expression and goblet cells hyperplasia. [21]
Regulation
The E-cadherin can regulate the SCCA production in the squamous cell carcinoma of the uterin cervix. E-cadherins are transmembrane proteins, they have a role in cell adhesion because they are able to form adherens junctions. They have to bind a Ca++ ion to work. Using an anti-E-cadherin antibody induces the dissociation of the cervical squamous cell carcinoma. It also induces a decrease of SCCA in the cytosol and SCCA m RNA. Besides the phosphatidyl inositol 3 kinase is a mediator of E-cadherin. The E-cadherin mediates cell-cell adhesion and maintains SCCA production thanks to phosphatidyl inositol 3 kinase in squamous cell carcinoma.[22]
References
- ↑ Martz, E. Book review of Introduction to protein science—architecture, function, and genomics: Lesk, Arthur M. Biochem. Mol. Biol. Educ. 33:144-5 (2006). DOI :10.1007/978-1-4612-0401-5_21#page-1
- ↑ Suminami Y, Nawata S, Kato H. Biological role of SCC antigen. Tumour Biol. 1998;19(6):488-93. PMID:9817978
- ↑ JBC Papers in Press. Published on July 2, 2001 as Manuscript R100016200 THE SERPINS ARE AN EXPANDING SUPERFAMILY OF
STRUCTURALLY SIMILAR BUT FUNCTIONALLY DIVERSE PROTEINShttp://www.jbc.org/content/early/2001/07/02/jbc.R100016200.full.pdf DOI : 2001/07/02/jbc.R100016200.full.pdf
- ↑ Zheng B, Matoba Y, Kumagai T, Katagiri C, Hibino T, Sugiyama M. Crystal structure of SCCA1 and insight about the interaction with JNK1. Biochem Biophys Res Commun. 2009 Feb 27;380(1):143-7. Epub 2009 Jan 21. PMID:19166818 doi:S0006-291X(09)00095-3
- ↑ Gary A. Silverman1*, Phillip I. Bird2
, Robin W. Carrell3
, Frank C. Church4
, Paul B. Coughlin5
, Peter G.W. Gettins6
,
James A Irving2
, David A. Lomas3
, Cliff J. Luke1
, Richard W. Moyer7
, Philip A. Pemberton8
, Eileen RemoldO'Donnell9
, Guy S. Salvesen10, James Travis11 and James C. Whisstock, THE SERPINS ARE AN EXPANDING SUPERFAMILY OF
STRUCTURALLY SIMILAR BUT FUNCTIONALLY DIVERSE PROTEINS, http://www.jbc.org/content/early/2001/07/02/jbc.R100016200.full.pdf DOI : 2001/07/02/jbc.R100016200.full.pdf
- ↑ PDB, Crystal structure of human squamous cell carcinoma antigen 1 http://www.rcsb.org/pdb/explore/remediatedSequence.do?structureId=2ZV6&bionumber=1 DOI : pdb/explore/remediatedSequence.do?structureId=2ZV6&bionumber=1
- ↑ PMID : PMC24842
- ↑ James C Whisstocka, 2, Richard Skinnera, 2, Robin W Carrella, Arthur M Leska, Conformational changes in serpins: I. the native and cleaved conformations of α1-antitrypsin1, http://www.sciencedirect.com/science/article/pii/S0022283699935209 DOI:pii/S0022283699935209
- ↑ M. S. J. Mangan, D. Kaiserman & P. I. Bird, The role of serpins in vertebrate immunity
Department of Biochemistry and Molecular Biology, Monash University, Clayton, Victoria, Australiahttp://onlinelibrary.wiley.com/doi/10.1111/j.1399-0039.2008.01059.x/pdf DOI : 10.1111/j.1399-0039.2008.01059.x/pdf
- ↑ James A. Irving1, Robert N. Pike1, Arthur M. Lesk2, and James C. Whisstock1, Phylogeny of the Serpin Superfamily: Implications of Patterns of Amino Acid Conservation for Structure and Function, http://genome.cshlp.org/content/10/12/1845 DOI : content/10/12/1845
- ↑ Sílvia Gomes, Patrícia I. Marques,Rune Matthiesen, Susana Seixas, Adaptive Evolution and Divergence of SERPINB3: A Young Duplicate in Great Apes,http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0104935 DOI : 10.1371%2Fjournal.pone.0104935<ref>
This move induce a protease distortion into a conformation, in which the acyl enzyme intermediate is hydrolysed extremely slowly. The active site of the enzyme would be expected to break.The protease thus remains covalently attached to the target protease and is thereby inhibited.
Consequently, the serpin has to be cleaved to inhibit the target protases. Thus, SCCA1 is irreversible protease inhibitor.
<ref>J. A. HUNTINGTON,Department of Haematology, Cambridge Institute for Medical Research, University of Cambridge, Cambridge, UK, Serpin structure, function and dysfunction,
http://onlinelibrary.wiley.com/doi/10.1111/j.1538-7836.2011.04360.x/pdf DOI : 10.1111/j.1538-7836.2011.04360.x/pdf</li>
<li id="cite_note-11">[[#cite_ref-11|↑]] L. P. Kerbrat, Que faire des marqueurs tumoraux, Centre Eugène Marquis, Université de Rennes 1 [https://facmed.univ-rennes1.fr/wkf/stock/RENNES20110504094607cpiszkormarqueurs_tumoraux.coursDCEM1-02-2011.pdf DOI : stock/RENNES20110504094607cpiszkormarqueurs_tumoraux.coursDCEM1-02-2011.pdf]</li>
<li id="cite_note-12">[[#cite_ref-12|↑]] Upham J, Campbell B. Utility of squamous cell carcinoma antigen (SCC Ag) as a tumour marker in pulmonary malignancy. Respir Med. 1992 May;86(3):201-3. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/1620906 1620906] </li>
<li id="cite_note-13">[[#cite_ref-13|↑]] Les marqueurs tumoraux Tableau d’aide à la description des principaux marqueurs tumoraux, Ketterhill laboratoires d’analyses médicales [http://www.llam.lu/fileadmin/media/newsletter/Marqueurs_Tum.pdf DOI : newsletter/Marqueurs_Tum.pdf] </li>
<li id="cite_note-14">[[#cite_ref-14|↑]] Uemura Y, Pak SC, Luke C, Cataltepe S, Tsu C, Schick C, Kamachi Y, Pomeroy SL, Perlmutter DH, Silverman GA. Circulating serpin tumor markers SCCA1 and SCCA2 are not actively secreted but reside in the cytosol of squamous carcinoma cells. Int J Cancer. 2000 Jul 20;89(4):368-77. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/10956412 10956412] </li>
<li id="cite_note-15">[[#cite_ref-15|↑]] Micke O, Prott FJ, Schäfer U, Tangerding S, Pötter R, Willich N.The impact of squamous cell carcinoma (SCC) antigen in the follow-up after radiotherapy in patients with cervical cancer. Anticancer Res 2000 ; 20 : 5113-5115. National Academy of Clinical Biochemistry.Guidelines for the Use of Tumor Markers in cervical cancer.[//www.nacb.org/lmpg/tumor/chp3j_cervical.d DOI : tumor/chp3j_cervical.d]</li>
<li id="cite_note-16">[[#cite_ref-16|↑]] Penelope L. Moore‡, Sarah Ong, and Tim J. Harrison§, Squamous Cell Carcinoma Antigen 1-mediated Binding of Hepatitis B Virus to Hepatocytes Does Not Involve the Hepatic Serpin Clearance System*[http://www.jbc.org/content/278/47/46709.full DOI 47/46709.full]</li>
<li id="cite_note-17">[[#cite_ref-17|↑]] Chika Katagiri, Jotaro Nakanishi, Kuniko Kadoya, and Toshihiko Hibino, Serpin squamous cell carcinoma antigen inhibits
UV-induced apoptosis via suppression of c-JUN
NH2-terminal kinase
http://jcb.rupress.org/content/172/7/983.full.pdf+html DOI : 172/7/983.full.pdf+html </li>
<li id="cite_note-18">[[#cite_ref-18|↑]] PMID : 19166818<ref>
===Receptor (LPR) interaction===
The serpin and serpin-protease complexes are able to bind the low density lipoprotein receptor-related protein (LPR). This binding allow to clear serpin- complexes from blood circulation. The ligand bind a clusters of rich cysteine residues. No differences were noticed between native and cleaved serpin. The binding between serpin and enzyme such as protease may increase the affinity of the complexe for LPR <ref>PMID: PMC2709341</li>
<li id="cite_note-19">[[#cite_ref-19|↑]] Santé médecine, Hyperplasie définition[http://sante-medecine.commentcamarche.net/faq/13479-hyperplasie-definition DOI : faq/13479-hyperplasie-definition]</li>
<li id="cite_note-20">[[#cite_ref-20|↑]] Morse GD, Holdsworth MT, Venuto RC, Gerbasi J, Walshe JJ. Pharmacokinetics and clinical tolerance of intravenous and oral cyclosporine in the immediate postoperative period. Clin Pharmacol Ther. 1988 Dec;44(6):654-64. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/3058372 3058372] </li>
<li id="cite_note-21">[[#cite_ref-21|↑]] Hirakawa H, Nawata S, Sueoka K, Murakami A, Takeda O, Numa F, Kato H, Sugino N. Regulation of squamous cell carcinoma antigen production by E-cadherin mediated cell-cell adhesion in squamous cell carcinoma cell line. Oncol Rep. 2004 Feb;11(2):415-9. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/14719077 14719077] </li></ol></ref>
Anything in this section will appear adjacent to the 3D structure and will be scrollable.


