We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 967
From Proteopedia
(Difference between revisions)
| Line 49: | Line 49: | ||
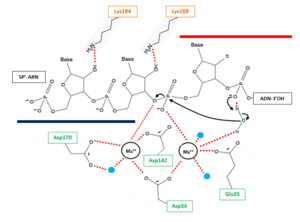
The hydrolysis can be decomposed in 3 steps: | The hydrolysis can be decomposed in 3 steps: | ||
* '''1''' : Deprotonation of a water molecule coordinated to the metal MB++ to form a nucleophile OH- ion. This hydroxide ion will then be properly oriented for an in-line nucleophilic attack of the target phosphate. The deprotonation mechanism has not been elucidated yet but two hypothesis can explain this step. According the first one, the metal MB++ might be responsible for the generation of water nucleophile. The other one involve a participation of the pro-R oxygen of the phosphate immediately to the 3’ side of the scissile bond, which is likely to serve as a general base for deprotonation (with transfer of the H+ to the solvent).This pro-R oxygen is also thought to play a role in the proper orientation of the hydroxide ion. | * '''1''' : Deprotonation of a water molecule coordinated to the metal MB++ to form a nucleophile OH- ion. This hydroxide ion will then be properly oriented for an in-line nucleophilic attack of the target phosphate. The deprotonation mechanism has not been elucidated yet but two hypothesis can explain this step. According the first one, the metal MB++ might be responsible for the generation of water nucleophile. The other one involve a participation of the pro-R oxygen of the phosphate immediately to the 3’ side of the scissile bond, which is likely to serve as a general base for deprotonation (with transfer of the H+ to the solvent).This pro-R oxygen is also thought to play a role in the proper orientation of the hydroxide ion. | ||
| - | * '''2''' : In line attack by hydroxide ion of the target phosphate. During this step, a pentacovalent phosphate (transition state) is formed and stabilized by metal MA++. The metal MA++ interacts with both the nonbridging and 3' bridging oxygen | + | * '''2''' : In line attack by hydroxide ion of the target phosphate. During this step, a pentacovalent phosphate (transition state) is formed and stabilized by metal MA++. The metal MA++ interacts with both the nonbridging and 3' bridging oxygen and the strain induced by this double briding is thought to accelerate the hydrolysis rate. This step release a 3’ oxyanion stabilized by metal MA++ acting as a Lewis acid. |
* '''3''' : The cleaved phosphate cannot simultaneously coordinate the two metal ions anymore, and likely one of the metal ions leave the active site which triggers a release of cleave product. | * '''3''' : The cleaved phosphate cannot simultaneously coordinate the two metal ions anymore, and likely one of the metal ions leave the active site which triggers a release of cleave product. | ||
Revision as of 09:55, 10 January 2015
| This Sandbox is Reserved from 15/11/2014, through 15/05/2015 for use in the course "Biomolecule" taught by Bruno Kieffer at the Strasbourg University. This reservation includes Sandbox Reserved 951 through Sandbox Reserved 975. |
To get started:
More help: Help:Editing |
Structure of the Mouse RNase H2 Complex
| |||||||||||
References
- ↑ http://genome-euro.ucsc.edu/cgi-bin/hgTracks?clade=mammal&org=Mouse&db=mm10&position=RnaseH2&hgt.positionInput=RnaseH2&hgt.suggestTrack=knownGene&Submit=submit&hgsid=201143152_yP1Xd4bMnHS7DV0d3VcqpDSxzzuQ&pix=1563
- ↑ 2.0 2.1 Rychlik, Monika P., Hyongi Chon, Susana M. Cerritelli, Paulina Klimek, Robert J. Crouch, and Marcin Nowotny. “Crystal Structures of RNase H2 in Complex with Nucleic Acid Reveal the Mechanism of RNA-DNA Junction Recognition and Cleavage.” Molecular Cell 40, no. 4 (November 24, 2010): 658–70. doi:10.1016/j.molcel.2010.11.001.
- ↑ Sparks, Justin L., Hyongi Chon, Susana M. Cerritelli, Thomas A. Kunkel, Erik Johansson, Robert J. Crouch, and Peter M. Burgers. “RNase H2-Initiated Ribonucleotide Excision Repair.” Molecular Cell 47, no. 6 (September 28, 2012): 980–86. doi:10.1016/j.molcel.2012.06.035.
- ↑ 4.0 4.1 4.2 Bubeck, Doryen, Martin A. M. Reijns, Stephen C. Graham, Katy R. Astell, E. Yvonne Jones, and Andrew P. Jackson. “PCNA Directs Type 2 RNase H Activity on DNA Replication and Repair Substrates.” Nucleic Acids Research 39, no. 9 (May 2011): 3652–66. doi:10.1093/nar/gkq980.
- ↑ Shaban, Nadine M., Scott Harvey, Fred W. Perrino, and Thomas Hollis. “The Structure of the Mammalian RNase H2 Complex Provides Insight into RNA•DNA Hybrid Processing to Prevent Immune Dysfunction.” Journal of Biological Chemistry 285, no. 6 (February 5, 2010): 3617–24. doi:10.1074/jbc.M109.059048.
- ↑ http://www.uniprot.org/uniprot/Q9CWY8
- ↑ http://www.uniprot.org/uniprot/Q80ZV0
- ↑ http://www.uniprot.org/uniprot/Q9CQ18
- ↑ Nicholson, Allen W. Ribonucleases. Springer Science & Business Media, 2011.
Proteopedia page contributors and editors
Anaïs Bourbigot & Valériane Keïta