Introduction
Antimicrobial Peptides (AMPs) are short peptides that consist of 10-50 amino acids, and were found to have antimicrobial influence on different kinds of bacteria and fungi. They are also called host defense peptides (HDPs) or defensins,Since they have different functions in their host.
AMPs are produced by Eukaryotes, as part of their defence mechanism from bacteria. They defend their host from bacteria, and also have physiological functions such as inflammation and wound healing (Wimley 2010)
Even though they have similar functions,AMPs lack any specific consensus amino acid sequences that are associated with biological activity.
History
The first AMP was discovered by Alexander Fleming about 90 years ago. He recognized the presence of a soluble antimicrobial substance produced by human nasal secretions from a patient suffering from acute coryza. He called it Lysozyme as it performed lysis to bacterial cells.
Subsequently, he found lysozyme antibacterial activity in various human physiological fluids and tissues of animals, as well as egg whites.
Since then, various natural examples for AMPs were found in all classes of organisms: animals including humans, invertebrate animals, plants, and fungi.
So far more than 1,200 types of peptides with antimicrobial activity have been isolated from various cells and tissues.
For a partial list of these, see the Antimicrobial Peptide Database[1]
Clasiffication
Antimicrobial peptides are divided into subgroups on the basis of their amino acid composition and structure (ref nature review).
(1) Anionic peptides
(1) Anionic peptides - Small anionic peptides rich in glutamic and aspartic acids from sheep, cattle and humans - they present in surfactant extracts, bronchoalveolar lavage fluid and airway epithelial cells. They are produced in mM concentrations, require zinc as a cofactor for antimicrobial activity and are active against both Gram positive and Gram-negative bacteria.
• Maximin H5 from amphibians.
• As example we can see Dermcidin peptide from human source: you can see here the . The aspartic acid is colored red and the glutamic acid is colored blue .
(2) Linear cationic α-helical peptides
(2) Linear cationic α-helical peptides - contains ~290 cationic peptides, which are short (contain <40 amino acid residues), lack
cysteine residues and sometimes have a hinge or ‘kink’ in the middle
In aqueous solutions many of these peptides are disordered, but in the presence of trifluoroethanol,sodium dodecyl sulphate (SDS) micelles, phospholipid vesicles and liposomes, or Lipid A, all or part of the molecule is converted to an α-helix.
It has been observed for buforin II, and LL-37, that the extent of α-helicity correlates with the antibacterial activity against both Gram-positive and Gram-negative bacteria — increased α-helical content correlates with stronger antimicrobial activitie
• Cecropins (A), andropin,moricin, ceratotoxin and melittin from insects.
• Cecropin P1 from Ascaris nematodes148.
• Magainin (2) (Magainin 2 in proteopedia), dermaseptin, bombinin, brevinin-1, esculentins and buforin II from
amphibians.
• Pleurocidin from skin mucous secretions of the winter flounder.
• Seminalplasmin,BMAP,SMAP (SMAP29,ovispirin), PMAP from cattle, sheep and pigs.
• CAP18 from rabbits.
• LL37 from human - In water, it exhibits a circular dichroism (CD) spectrum that is consistent with a disordered structure.However, in 15 mM HCO3–, SO42– or CF3CO2–, the peptide adopts a helical structure.
(3) Cationic peptides enriched for specific amino acids
(3) Cationic peptides enriched for specific amino acids - contains ~44 cationic peptides that are rich in certain amino acids. These peptides lack cysteine residues and are linear, although some can form extended coilsץ
• Proline-containing peptides include abaecin from honeybees.
• Proline- and arginine-containing peptides include apidaecins from honeybees; drosocin from Drosophila; pyrrhocoricin from the European sap-sucking bug; bactenecins from cattle (Bac7), sheep, and goats; and PR-39 from pigs.
• Proline- and phenylalanine-containing peptides include prophenin from pigs.
• Glycine-containing peptides include hymenoptaecin from honeybees.
• Glycine- and proline-containing peptides include coleoptericin and holotricin from beetles28.
• Tryptophan-containing peptides include from cattle. Indolicidn is consists of 13 amino acids include , and .
• Small histidine-rich salivary polypeptides, including the histatins from man and some higher primates116.
(4) Anionic and cationic peptides that contain cysteine and form disulphide bonds
(4) Anionic and cationic peptides that contain cysteine and form disulphide bonds - this group has ~380 members, contain cysteine residues and form disulphide bonds and stable β-sheets.
A diverse family of defensins (defensin in proteopeia) is belong to this group.
There are ~55 α-defensins, which include human neutrophil peptides (HNPs) and cryptdins and comprise
29–35 amino acid residues, including six cysteines that are linked by three intramolecular disulphide bonds.
There are ~90 β-defensins from both humans (HBDs) and animals that comprise 36–42 amino acid residues including six cysteines that are linked by three intramolecular disulphide bonds.
In addition, there are ~54 arthropod (insect) defensins, ~58 plant defensins and a rhesus θ-defensin (RTD-1), which is an 18-residue peptide that forms a circular molecule that is crosslinked by three disulphide bonds. SPAG11/isoform HE2C is an atypical anionic β-defensin-like peptide.
• Peptides with 1 disulphide bond include brevinins.
• Peptides with 2 disulphide bonds include from pigs and tachyplesins from horseshoe crabs.
is from porcine leukocytes (which comprises 16 amino acid residues, including
four cysteines that are linked by two intramolecular disulphide bonds).
• Peptides with 3 disulphide bonds include α-defensins from humans (HNP-1,HNP-2,cryptidins), rabbits (NP-1) and rats; β-defensins from humans (HBD1, DEFB118),cattle,mice, rats, pigs, goats and poultry12; and rhesus θ-defensin (RTD-1) from the rhesus monkey.
• Insect defensins (defensin A).
• SPAG11/isoform HE2C, an atypical anionic β-defensin.
• Peptides with >3 disulphide bonds include plant antifungal defensins and in fruit flies. Drosomycin is the first antifungal protein characterized recently among the broad family of inducible peptides and proteins produced by insects to respond to bacterial or septic injuries. It contains .
(5) Anionic and cationic peptide fragments of larger proteins
(5) Anionic and cationic peptide fragments of larger proteins - these fragments have antimicrobial activity and are similar in composition and structure to the antimicrobial peptides described above. However, their role in innate immunity is not yet clear.
• Lactoferricin from lactoferrin (human lactoferrin in proteopedia).
• Casocidin I from human casein.
• Antimicrobial domains from bovine α-lactalbumin, human haemoglobin, lysozyme (Hen Egg-White (HEW) Lysozyme in proteopedia) and ovalbumin.
Structural highlights
AMPs shared the abillity to attach membranes. Amino acids composition as well as the structure allow each of them to attach the microorganism's membrane.
Primary sequence
AMPs are rich with hydrophibic (Ala, Val, Ile, Leu, Met, Phe, Tyr, Trp) and Possitively charged (Lys, Arg) Amino Acids, which seems to allow them to bind into membranes. , is a peptide from porcine leukocytes and it's sequence is rich with and .
Secondary structure
AMPs structure allows them to interact with negatively charged phospholipid head groups of microbial membranes, resulting in pore formation (or other mechanism) on the bacterial membrane .
Although AMPs have the same effect on the cell mambrane, they do not seem to have the same structure. we can find a big variety of structures among familiar AMPs.
(1) Some have helical structures, for some of the peptide sequence, such as ,, or a helical structure throughout the whole peptide, such as . this peptide was found on a frog's skin. you can see the page about Magainin2 here : Magainin 2.
(2) Beta-sheet structures:
, A cyclic antimicrobial defencin from Rhesus Macaque leukocytes,has a beta sheet structure, rich with disulfide bonds that strengthen the beta-sheet structure. this peptide is 55% beta sheet (4 strands; 10 residues).
from porcine leukocytes, NMR, has a hairpin shape
63% beta sheet, 2 strands, 12 residues
Protegrins are a family of arginine - and cysteine rich cationic peptides
(c) and some have combined structures, like
Suggested Mechanisms
For killing the bacteria, AMPs must first attracted to bacterial surfaces. The attraction is electrostatic bonding between anionic or cationic peptides and structures on the bacteria's membrane. Studies on membrane models contained vary compositions of charged phospholipids proved this mechanism of attraction. But, bacteria membranes are more complex than mebrane models. Gram negative - Cationic AMPS suggested to be attracted to the net negative charge on the puter mmebrane (phospholipids to phosphate groupf of lipopolysacharides). Gram positive - AMPSs suggested to be attracted to the trichoic acids ob the surface.
Now, the peptides are attached to the cell. However they have to tranverse the lipopolysacharides or extracellular polysacharides so they can contact and act on the outer membrane. the specific mechanism of this step is unclear so far.
The way how different antimicrobial peptides insert and act on the membrane goal appears to be different and depends on the peptide, the membrane and the peptide/lipid ratio.
At low peptide/lipid ratios, peptides are oriented parralel to the lipid bilayer,
As the peptide/lipid ration increases, peptides are oriented prependicular to the membrane and insert inside it, forming transmembranes pores.
There are a few suggested machanisms of how AMPs work (William C. Wimley, ACS CHEMICAL BIOLOGY, 2010).
They can be divided into two:
(A) Transmembrane Pore Models of AMP Membrane Activity and (B) Nonpore Models of AMP Activity
In the Transmembrane Pore Models, it is suggested that AMPs form many pores in the mambrane, so that it cannot hold it's content anymore.
tha transmembrane pore mechanism has 2 main models:
1- barrel stave pore model ,that claims peptides interact laterally with one another to form a specific structure enclosing a water-filled channel, much like a protein ion channel.
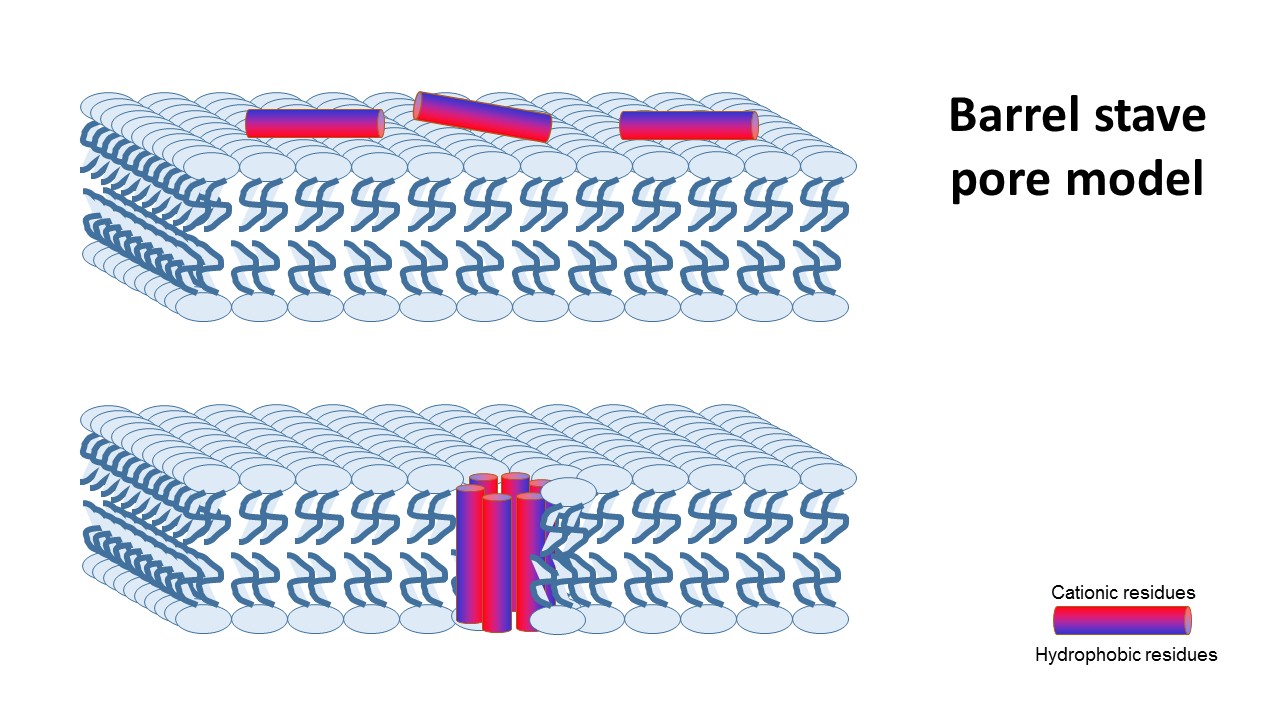
2- toroidal pore model, that claims specific peptide–peptide interactions are not present, and instead, single peptides are bound to the membrane’s phospholipids and disturbe it’s structure..
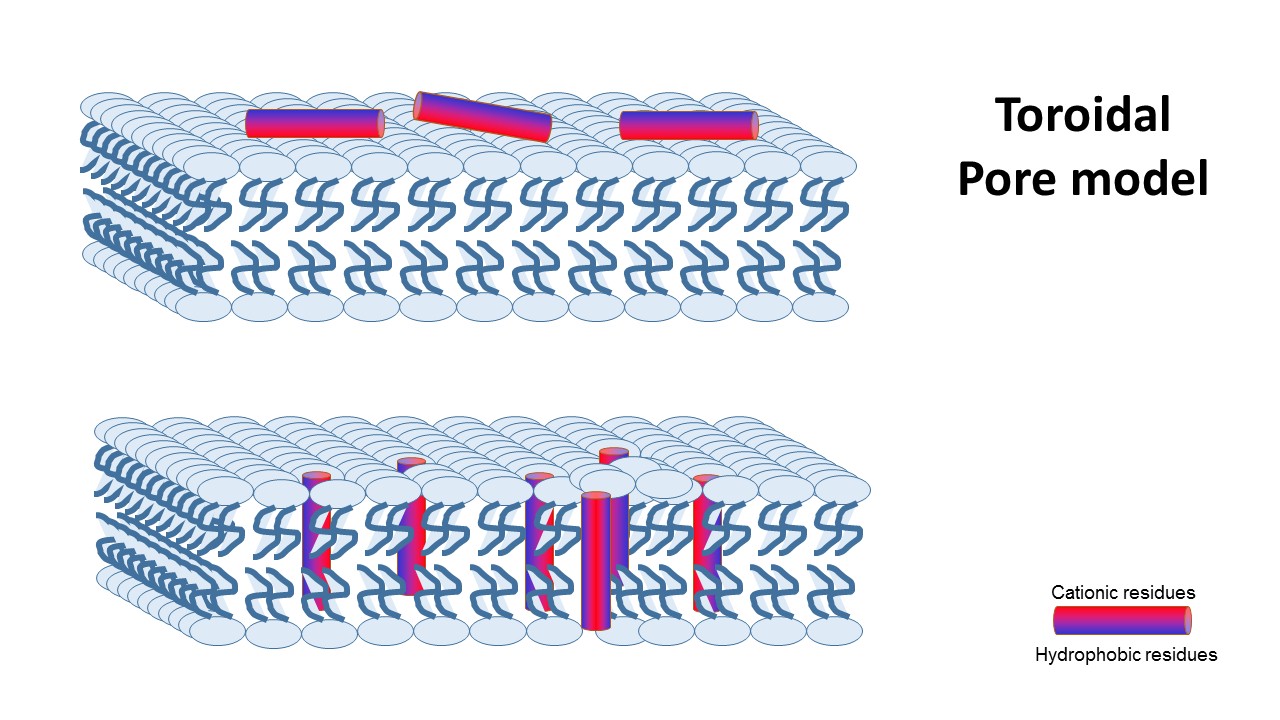
the Nonepore model claims peptides bind to the membrane until it collapses. It is devided into 2 main mechanisms:
1- The carpet model. In this model, antimicrobial peptides accumulate on the membrane surface with an orientation that is parallel to the membrane.When peptide concentration has reached a critical level permeabilization occurs via global bilayer destabilization.
2- detergent model- collapse of membrane integrity, observed with some AMPs at high peptide concentration.
Image:Detregent model.JPG
Relevance


