Sandbox Reserved 1086
From Proteopedia
| Line 17: | Line 17: | ||
== Exploring the Structure == | == Exploring the Structure == | ||
[[Image: Screen_Shot_2015-04-22_at_12.53.12_PM.png|thumb|right|220px|Fig.1. ''Greek key'' fold]] | [[Image: Screen_Shot_2015-04-22_at_12.53.12_PM.png|thumb|right|220px|Fig.1. ''Greek key'' fold]] | ||
| - | [[Image: | + | [[Image: CALCIUM_COORD.png|thumb|right|220px|Fig.2. Different calcium coordinations]] |
[[Image: Screen_Shot_2015-04-20_at_5.38.13_PM.png|thumb|right|220px|Fig.3. First directional bending between EC1 and EC2]] | [[Image: Screen_Shot_2015-04-20_at_5.38.13_PM.png|thumb|right|220px|Fig.3. First directional bending between EC1 and EC2]] | ||
[[Image: Screen_Shot_2015-04-20_at_5.27.27_PM.png|thumb|right|220px|Fig.4. Second directional bending between EC2 and EC3]] | [[Image: Screen_Shot_2015-04-20_at_5.27.27_PM.png|thumb|right|220px|Fig.4. Second directional bending between EC2 and EC3]] | ||
Revision as of 12:08, 22 April 2015
| This Sandbox is Reserved from 15/04/2015, through 15/06/2015 for use in the course "Protein structure, function and folding" taught by Taru Meri at the University of Helsinki. This reservation includes Sandbox Reserved 1081 through Sandbox Reserved 1090. |
To get started:
More help: Help:Editing |
Contents |
C-cadherin
| |||||||||||
Exploring the Structure
The extracellular moiety of the protein consists of five ectodomains () alternated by four . Each EC shares the same greek key fold, characteristic of immunoglobulin (Fig.1). The interfaces between the EC are calcium binding sites coordinating three ions each through the highly conserved motifs Asp-X-Asp, Leu-Asp-Arg-Glu and Asp-X-Asn-Asp-Asn. The binding of calcium is thought to confer rigidity as well as the classical curved shape to the structure and this is why it´s suggested to be a triggering if not even compulsory step to the adhesion process.
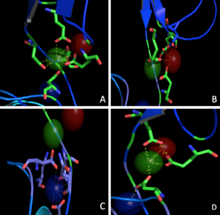
Calcium Coordination
Three calcium ions are differently coordinated in each interface between two neighbouring EC. Just one of these seems to be coordinated with the classical epta-coordination characteristic of the elements belonging to the first and second group (Figx) while the others are bound without any particular geometry (Figy/z). Nevertheless, the presence of common coordinating residues (Glu) links these ions one to another forming a net of strong interactions (Fig.r).
Molecular Geometry
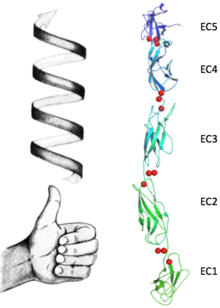
Upon the calcium binding each EC contracts to the neighbour assuming a defined position which leads to the bended structure of the protein. These conformations changes in a sequel of ions form the unique final conformation of this protein. In Fig.2 the bending between EC1 and EC2 is shown in just one dimension, making clear the angle between the two main axes (approximately 22.5°). Fig.3, represents the bending between EC2 and EC3 while concealing this property between first and second EC. Because of this concealing due to the change in the observed direction, the change of position of EC3 respect EC2 can be no longer addressed the same direction of the bending between EC1 and EC2 but to a different one. The occurrence of bendings in (at least) two different directions confers a torsional behaviour which goes through all the EC. These particular fashion contributes to the complexity of the protein´s ternary structure, shaping it into a bended left handed helix (Fig.4).
Adhesion Models
This conformation (called S) might play a key role in tackling the pull forces within the desmosomal plaque. In fact, two cadherins that take contact from two different surfaces are going to perpetuate the left handed shape to the quaternary structure. As consequence the dimer unit connecting the two cells will resist to the stretching force with the same features of a spring (Fig.5). The S shape conformation shown in this model is not the only one observed.
Glycosylation
C cadherin is a N/O-glycosilated protein. It counts 12 sugars attached all over the structure to Thr-Asn residues but mainly to EC3 and EC4. Their role in the adhesion process in unknown and not suspected. Being transmembrane proteins, the presence of these sugars might be involved in the immune self-recognition.
Structural highlights
| 1q5a is a 2 chain structure with sequence from Mus musculus. The March 2008 RCSB PDB Molecule of the Month feature on Cadherin by David S. Goodsell is 10.2210/rcsb_pdb/mom_2008_3. Full crystallographic information is available from OCA.
This is a view if the molecule by groups, and the of the protein.
For a guided tour on the structure components use FirstGlance. | |
| Ligands: | , , |
| Related: | 1l3w, 1q55, 1q5b, 1q5c |
| Resources: | FirstGlance, OCA, RCSB, PDBsum |