We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1083
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
Four molecules of AcrA (45-312 residues) in asymmetric unit of the crystal pack as an apparent <scene name='69/699996/Acra-dimer_of_dimers/1'>dimer of dimers</scene>. Each monomers are labeled as A (in cyan), B (in orange), C (in green) and D (in yellow). A, B / C, D are related to one another by approximate <scene name='69/699996/Dyad_symmetry/3'>dyad symmetry</scene>. Each set of dimers are related to one another by approximate <scene name='69/699996/2_fold_symmetry/2'>2 fold axis</scene> <ref>PMID:16531241</ref>. | Four molecules of AcrA (45-312 residues) in asymmetric unit of the crystal pack as an apparent <scene name='69/699996/Acra-dimer_of_dimers/1'>dimer of dimers</scene>. Each monomers are labeled as A (in cyan), B (in orange), C (in green) and D (in yellow). A, B / C, D are related to one another by approximate <scene name='69/699996/Dyad_symmetry/3'>dyad symmetry</scene>. Each set of dimers are related to one another by approximate <scene name='69/699996/2_fold_symmetry/2'>2 fold axis</scene> <ref>PMID:16531241</ref>. | ||
| - | Each <scene name='69/699996/Monomer/1'>monomer</scene> is a sickle shaped molecule comprising three domains viz. <scene name='69/699996/Beta_barrel/1'>β-barrel domain</scene>, <scene name='69/699996/Lipoyl_domain/2'>lipoyl domain</scene>, and coiled coil <scene name='69/699996/Hairpin_domain/1'>α-helical hairpin domain</scene>. β-barrel domain comprises six anti-parallel β-sheets and a short α-helix. Lipoyl domain is present in the central part of the AcrA monomer made up of two half motifs interrupted by an α-helical hairpin. Each half of the lipoyl motif is homologous to each other and consist of four β-strands in the form of a β-sandwich. A conserved <scene name='69/699996/Lysine_residue/1'>lysine residue</scene> on the connecting loop of two half motifs serve as carrier of lipoyl or biotinyl co-factors. The coiled coil domain consists of five [http://en.wikipedia.org/wiki/Heptad_repeat heptad repeats] per helix. Two α-helices are packed together as a canonical [http://en.wikipedia.org/wiki/Knobs_into_holes_packing knobs-into-holes] by hydrophobic side chains in the <scene name='69/699996/Knob_in_hole/2'>a and d positions</scene> of the heptad repeats | + | Each <scene name='69/699996/Monomer/1'>monomer</scene> is a sickle shaped molecule comprising three domains viz. <scene name='69/699996/Beta_barrel/1'>β-barrel domain</scene>, <scene name='69/699996/Lipoyl_domain/2'>lipoyl domain</scene>, and coiled coil <scene name='69/699996/Hairpin_domain/1'>α-helical hairpin domain</scene>. β-barrel domain comprises six anti-parallel β-sheets and a short α-helix. Lipoyl domain is present in the central part of the AcrA monomer made up of two half motifs interrupted by an α-helical hairpin. Each half of the lipoyl motif is homologous to each other and consist of four β-strands in the form of a β-sandwich. A conserved <scene name='69/699996/Lysine_residue/1'>lysine residue</scene> on the connecting loop of two half motifs serve as carrier of lipoyl or biotinyl co-factors. The coiled coil domain consists of five [http://en.wikipedia.org/wiki/Heptad_repeat heptad repeats] per helix. Two α-helices are packed together as a canonical [http://en.wikipedia.org/wiki/Knobs_into_holes_packing knobs-into-holes] by hydrophobic side chains in the <scene name='69/699996/Knob_in_hole/2'>a and d positions</scene> of the heptad repeats <ref>PMID:10092468</ref> <ref>PMID:15507433</ref>. Crystal structure provide evidence for the flexibility of the hinge region between α-helical hairpin and lipoyl domain. The difference in hinge angle in case of B and C chain varies approximately by 15° overall and 21 Å at the loop located at the top of the hairpin. |
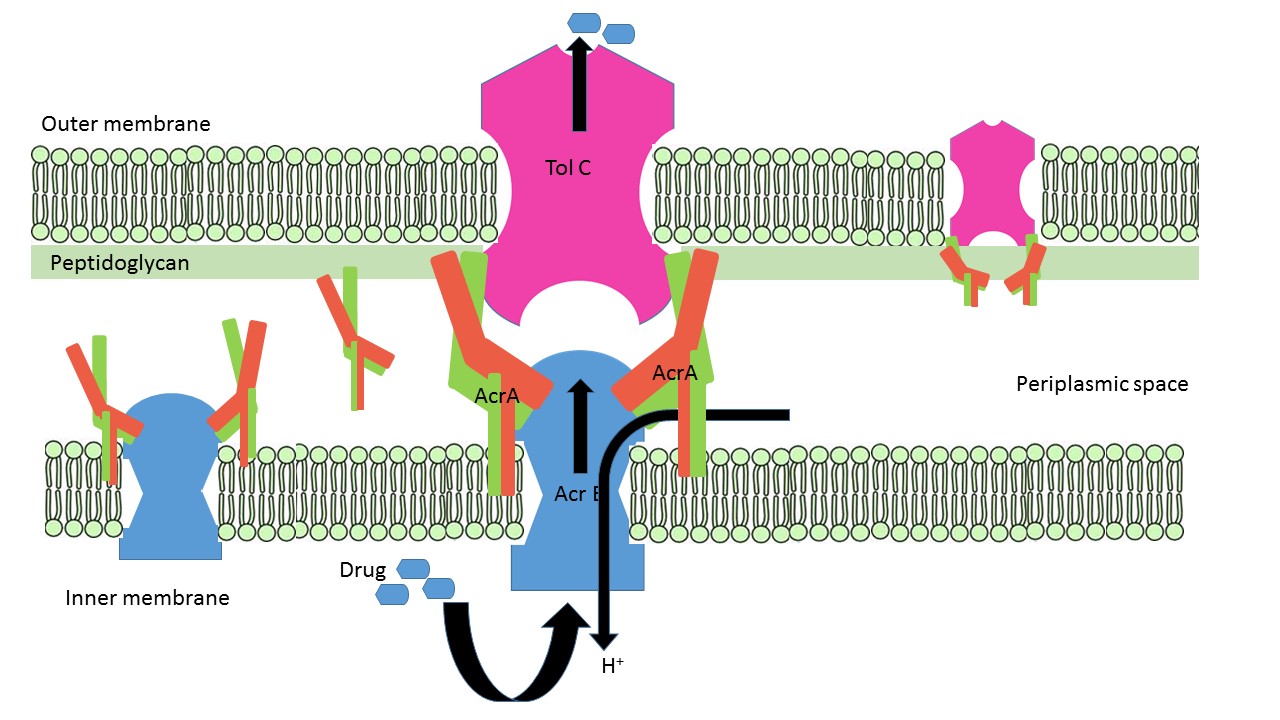
== Assembly within biological system == | == Assembly within biological system == | ||
Revision as of 06:54, 23 April 2015
| This Sandbox is Reserved from 15/04/2015, through 15/06/2015 for use in the course "Protein structure, function and folding" taught by Taru Meri at the University of Helsinki. This reservation includes Sandbox Reserved 1081 through Sandbox Reserved 1090. |
To get started:
More help: Help:Editing |
AcrA
| |||||||||||
References
- ↑ Putman M, van Veen HW, Konings WN. Molecular properties of bacterial multidrug transporters. Microbiol Mol Biol Rev. 2000 Dec;64(4):672-93. PMID:11104814
- ↑ Kumar A, Schweizer HP. Bacterial resistance to antibiotics: active efflux and reduced uptake. Adv Drug Deliv Rev. 2005 Jul 29;57(10):1486-513. PMID:15939505 doi:http://dx.doi.org/10.1016/j.addr.2005.04.004
- ↑ Mikolosko J, Bobyk K, Zgurskaya HI, Ghosh P. Conformational flexibility in the multidrug efflux system protein AcrA. Structure. 2006 Mar;14(3):577-87. PMID:16531241 doi:10.1016/j.str.2005.11.015
- ↑ Johnson JM, Church GM. Alignment and structure prediction of divergent protein families: periplasmic and outer membrane proteins of bacterial efflux pumps. J Mol Biol. 1999 Apr 2;287(3):695-715. PMID:10092468 doi:http://dx.doi.org/10.1006/jmbi.1999.2630
- ↑ Akama H, Kanemaki M, Yoshimura M, Tsukihara T, Kashiwagi T, Yoneyama H, Narita S, Nakagawa A, Nakae T. Crystal structure of the drug discharge outer membrane protein, OprM, of Pseudomonas aeruginosa: dual modes of membrane anchoring and occluded cavity end. J Biol Chem. 2004 Dec 17;279(51):52816-9. Epub 2004 Oct 26. PMID:15507433 doi:10.1074/jbc.C400445200