This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 1084
From Proteopedia
(Difference between revisions)
| Line 38: | Line 38: | ||
== Structural highlights == | == Structural highlights == | ||
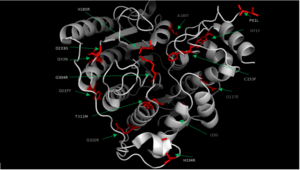
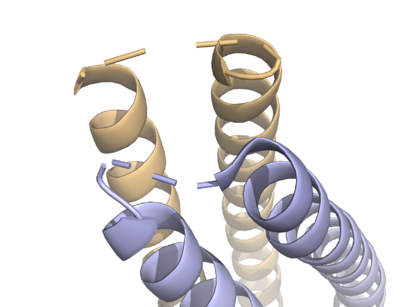
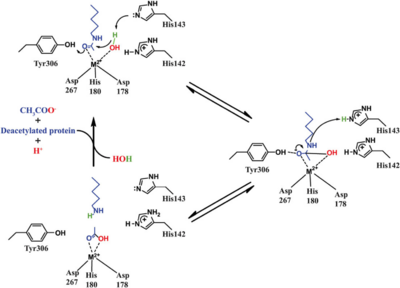
HDAC8 was the first among the HDAC isozymes whose crystal structure became available. The image shows the ribbon structure of <scene name='69/699997/Hdac8_saha/10'>HDAC8 bound with a canonical hydroxamate inhibitor, SAHA (Suberoylanilide Hydroxamic Acid)</scene>. HDAC8 contains a single α/β-deacetylase domain consisting of <scene name='69/699997/Hdac8_saha/11'>13 α-helices</scene> and an <scene name='69/699997/Hdac8_saha/13'>eight-stranded parallel β-sheet</scene>. [[Image:Loops.png|400px|left|thumb| '''Fig. 3''' The various loops associated with HDAC8.]]Multiple loops, which emanate from the protein core, play a significant role in maintaining the appropriate geometry of the catalytic pocket. The α/β-fold of the above kind was first observed in a metalloenzyme, [http://proteopedia.org/wiki/index.php/Arginase Arginase]. The active site of HDAC8 contains a tubular cavity leading to catalytic machinery at the end. The <scene name='69/699997/Hdac8_active_site_saha/18'>catalytic machinery is comprised of a Zn<sup>2+</sup> ion penta-coordinated</scene> with a square pyramidal geometry. The residues His 180, Asp 267 and Asp 178 occupy the three co-ordination sites, whereas the hydroxamate moiety of SAHA occupies the remaining two sites. In addition, the carbonyl oxygen of the hydroxamate moiety forms a hydrogen bond with Tyr 306. In the absence of any ligand (substrate/inhibitors), two water molecules are bound to the Zn<sup>2+</sup> ion. <scene name='69/699997/Dac8_active_site_haha/9'>The linker domain of the inhibitor interacts with the residues: Phe 152, Phe 208, His 180, Gly 151, and Met 274</scene>, which forms a <scene name='69/699997/Hdac8_tunnel/9'>hydrophobic tunnel</scene>. Notably, the above residues involved in the inhibitor/substrate binding, as well as the catalysis, have been found to be conserved during the course of evolution among class I HDACs. Notably, the carbonyl oxygen of the acetyl-lysine substrate forms a hydrogen bond with the hydroxyl moiety of Y306. | HDAC8 was the first among the HDAC isozymes whose crystal structure became available. The image shows the ribbon structure of <scene name='69/699997/Hdac8_saha/10'>HDAC8 bound with a canonical hydroxamate inhibitor, SAHA (Suberoylanilide Hydroxamic Acid)</scene>. HDAC8 contains a single α/β-deacetylase domain consisting of <scene name='69/699997/Hdac8_saha/11'>13 α-helices</scene> and an <scene name='69/699997/Hdac8_saha/13'>eight-stranded parallel β-sheet</scene>. [[Image:Loops.png|400px|left|thumb| '''Fig. 3''' The various loops associated with HDAC8.]]Multiple loops, which emanate from the protein core, play a significant role in maintaining the appropriate geometry of the catalytic pocket. The α/β-fold of the above kind was first observed in a metalloenzyme, [http://proteopedia.org/wiki/index.php/Arginase Arginase]. The active site of HDAC8 contains a tubular cavity leading to catalytic machinery at the end. The <scene name='69/699997/Hdac8_active_site_saha/18'>catalytic machinery is comprised of a Zn<sup>2+</sup> ion penta-coordinated</scene> with a square pyramidal geometry. The residues His 180, Asp 267 and Asp 178 occupy the three co-ordination sites, whereas the hydroxamate moiety of SAHA occupies the remaining two sites. In addition, the carbonyl oxygen of the hydroxamate moiety forms a hydrogen bond with Tyr 306. In the absence of any ligand (substrate/inhibitors), two water molecules are bound to the Zn<sup>2+</sup> ion. <scene name='69/699997/Dac8_active_site_haha/9'>The linker domain of the inhibitor interacts with the residues: Phe 152, Phe 208, His 180, Gly 151, and Met 274</scene>, which forms a <scene name='69/699997/Hdac8_tunnel/9'>hydrophobic tunnel</scene>. Notably, the above residues involved in the inhibitor/substrate binding, as well as the catalysis, have been found to be conserved during the course of evolution among class I HDACs. Notably, the carbonyl oxygen of the acetyl-lysine substrate forms a hydrogen bond with the hydroxyl moiety of Y306. | ||
| - | Aside from the catalytic Zn<sup>2+</sup> ion, the enzyme activity of HDAC8 is dependent on the presence of the monovalent ion, K<sup>+</sup>/Na<sup>+</sup> | + | Aside from the catalytic Zn<sup>2+</sup> ion, the enzyme activity of HDAC8 is dependent on the presence of the monovalent ion, K<sup>+</sup>/Na<sup>+</sup> <ref>Gantt, S. L., Joseph, C. G. & Fierke, C. A. Activation and Inhibition of Histone Deacetylase 8 by Monovalent Cations. J. Biol. Chem. 285, 6036–6043 (2010).</ref>. The crystal structure of HDAC8 shows the presence of two binding sites for K<sup>+</sup>/Na<sup>+</sup> <ref>Vannini, A. et al. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc. Natl. Acad. Sci. U.S.A. 101, 15064–15069 (2004)</ref>. The <scene name='69/699997/K1_site/4'>first K+/Na+ binding site (K1)</scene> is located in the vicinity of the enzyme catalytic machinery, and it is hexacoordinated (octahedral geometry) with His 180 (carbonyl oxygen of the main chain), Asp 178 (oxygen atom of the main chain and side chain), Leu 200 (carbonyl oxygen of the main chain), and Ser 199 (O). Notably, His 180 and Asp 178 are the common residues coordinated with both the catalytic Zn<sup>2+</sup> as well as the K<sup>+</sup>/Na<sup>+</sup> ion. The <scene name='69/699997/K2_site/4'>second binding site for K+/Na+ ion (K2)</scene> is located near the surface. It is hexacoordinated (octahedral geometry) with F189, T192, V195, Y225 as well as two water molecule. |
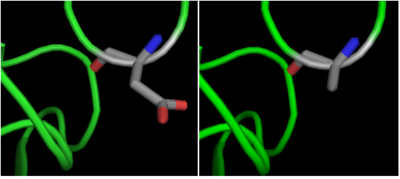
The crystal structure of <scene name='69/699997/Hdac8_binding_substrate/3'>HDAC8-substrate complex</scene> shows that aspartate residue <scene name='69/699997/Hdac8_d101/2'>(D101)</scene> is important in the substrate binding <ref>Vannini, A. et al. Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8–substrate complex. EMBO reports 8, 879–884 (2007).</ref>. The Asp 101 carboxylate moiety makes two consecutive hydrogen bonds with the backbone of the deacetylated peptide substrate. Mutation of Asp 101 to Ala inhibits HDAC8 activity. The Asp residue has been found to be strictly conserved among different HDAC isozymes. | The crystal structure of <scene name='69/699997/Hdac8_binding_substrate/3'>HDAC8-substrate complex</scene> shows that aspartate residue <scene name='69/699997/Hdac8_d101/2'>(D101)</scene> is important in the substrate binding <ref>Vannini, A. et al. Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8–substrate complex. EMBO reports 8, 879–884 (2007).</ref>. The Asp 101 carboxylate moiety makes two consecutive hydrogen bonds with the backbone of the deacetylated peptide substrate. Mutation of Asp 101 to Ala inhibits HDAC8 activity. The Asp residue has been found to be strictly conserved among different HDAC isozymes. | ||
Current revision
| This Sandbox is Reserved from 15/04/2015, through 15/06/2015 for use in the course "Protein structure, function and folding" taught by Taru Meri at the University of Helsinki. This reservation includes Sandbox Reserved 1081 through Sandbox Reserved 1090. |
To get started:
More help: Help:Editing |
Histone Deacetylase 8
| |||||||||||
References
- ↑ Ramakrishnan, V. Histone Structure and the Organization of the Nucleosome. Annual Review of Biophysics and Biomolecular Structure 26, 83–112 (1997).
- ↑ Waltregny, D. et al. Histone deacetylase HDAC8 associates with smooth muscle alpha-actin and is essential for smooth muscle cell contractility. FASEB J. 19, 966–968 (2005).
- ↑ Haberland, M., Mokalled, M. H., Montgomery, R. L. & Olson, E. N. Epigenetic control of skull morphogenesis by histone deacetylase 8. Genes Dev. 23, 1625–1630 (2009).
- ↑ Gräff, J. & Tsai, L.-H. The Potential of HDAC Inhibitors as Cognitive Enhancers. Annual Review of Pharmacology and Toxicology 53, 311–330 (2013).
- ↑ Blagosklonny, M. V. et al. Histone deacetylase inhibitors all induce p21 but differentially cause tubulin acetylation, mitotic arrest, and cytotoxicity. Mol. Cancer Ther. 1, 937–941 (2002).
- ↑ 6.0 6.1 6.2 Yan, W. et al. Histone deacetylase inhibitors suppress mutant p53 transcription via histone deacetylase 8. Oncogene 32, 599–609 (2013).
- ↑ Dokmanovic, M., Clarke, C. & Marks, P. A. Histone Deacetylase Inhibitors: Overview and Perspectives. Mol Cancer Res 5, 981–989 (2007).
- ↑ Bolden, J. E., Peart, M. J. & Johnstone, R. W. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov 5, 769–784 (2006).
- ↑ 9.0 9.1 Deardorff, M. A. et al. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature 489, 313–317 (2012).
- ↑ Ito, K. et al. Decreased histone deacetylase activity in chronic obstructive pulmonary disease. N. Engl. J. Med. 352, 1967–1976 (2005).
- ↑ Gantt, S. L., Joseph, C. G. & Fierke, C. A. Activation and Inhibition of Histone Deacetylase 8 by Monovalent Cations. J. Biol. Chem. 285, 6036–6043 (2010).
- ↑ Vannini, A. et al. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc. Natl. Acad. Sci. U.S.A. 101, 15064–15069 (2004)
- ↑ Vannini, A. et al. Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8–substrate complex. EMBO reports 8, 879–884 (2007).
- ↑ 14.0 14.1 Somoza, J. R. et al. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure 12, 1325–1334 (2004).