User:Wade Cook/Sandbox 1
From Proteopedia
| Line 7: | Line 7: | ||
== Function == | == Function == | ||
| - | The smallpox virus invades host cells by binding to specific receptors on the host cell membrane. The virus binds to host cell receptors via hemagglutinin antigens expressed on its outer surface. The exact mechanism of entry is not yet known. Upon invasion of a host cell, the smallpox virus replicates in the host cytoplasm, rather than the host nucleus, using many of it’s own enzymes to replicate. Replication begins once the virion has reached the cytoplasm of the host cell. First, a messenger RNA molecule is transcribed by RNA polymerase and related enzymes before the genome is uncoated. The first genes that are transcribed, known as early genes, code for proteins that facilitate the uncoating of the viral genome and initiate a second round of transcription of intermediate genes. The intermediate genes produce mRNA which are translated into proteins that allow the transcription of the late class of genes. The late class genes encode proteins which make up the structural and enzymatic components of a new virion. Further replication of this virus leads to the eventual shutdown of the host cell’s DNA, RNA, and protein synthesis, and causes changes to the cell’s architecture to allow the virus to use the host cell’s genetic machinery for reproduction. | + | The smallpox virus invades host cells by binding to specific receptors on the host cell membrane. The virus binds to host cell receptors via hemagglutinin antigens expressed on its outer surface. The exact mechanism of entry is not yet known. Upon invasion of a host cell, the smallpox virus replicates in the host cytoplasm, rather than the host nucleus, using many of it’s own enzymes to replicate. Replication begins once the virion has reached the cytoplasm of the host cell. First, a messenger RNA molecule is transcribed by RNA polymerase and related enzymes before the genome is uncoated. The first genes that are transcribed, known as early genes, code for proteins that facilitate the uncoating of the viral genome and initiate a second round of transcription of intermediate genes. The intermediate genes produce mRNA which are translated into proteins that allow the transcription of the late class of genes. The late class genes encode proteins which make up the structural and enzymatic components of a new virion. Further replication of this virus leads to the eventual shutdown of the host cell’s DNA, RNA, and protein synthesis, and causes changes to the cell’s architecture to allow the virus to use the host cell’s genetic machinery for reproduction (Berwald, 2004). |
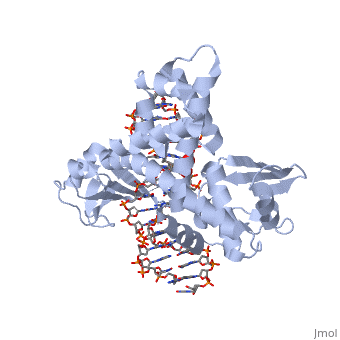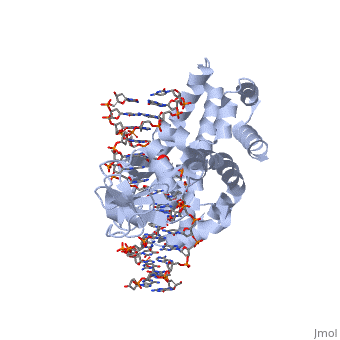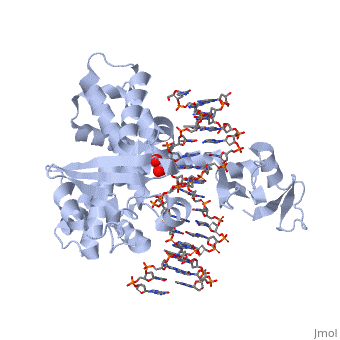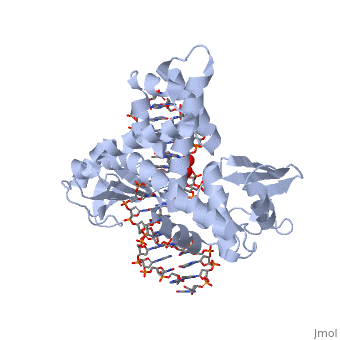
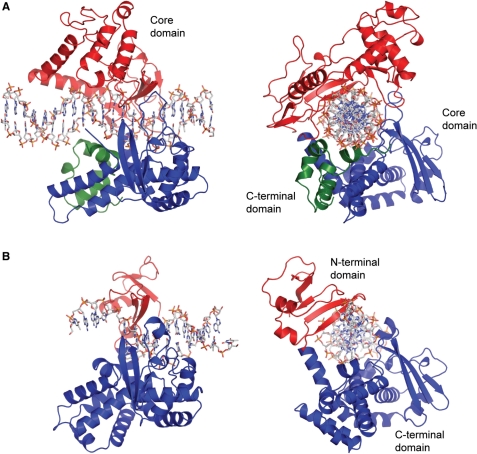
| - | One characteristic that makes the Variola virus so deadly is that it doesn’t require the host cell’s genetic machinery to begin replication. The reason for this is that the Variola virus codes for all the enzymes needed for its proliferation. One enzyme in particular, the type IB topoisomerase is responsible for unwinding the packaged, supercoiled, viral DNA to initiate viral replication. This is accomplished through a nick-joining mechanism of the double stranded DNA, in which a tyrosine nucleophile attacks a phosphodiester bond to form a 3’ phosphotyrosine linkage and releases a free 5’ hydroxyl group. This creates a nick to unwind supercoiling, which is then re-ligated to form relaxed DNA. Most mechanisms involving DNA including replication, transcription, and repair induce a certain degree of positive supercoiling. Tension due to the positive supercoiling buildup can be relieved by the enzymes topoisomerase. The variola topoisomerase 1B is an unusual topoisomerase. It is by far the smallest topoisomerase (33Kda) and only acts at specific sites that contain the sequence 5’-(T/C)CCTT-3’ in the variola genome. The topoisomerase 1B is made up of two domains that wrap around this recognition sequence forming a clamp around the DNA. The smaller amino terminal domain is responsible for interactions with the DNA sequence, whereas the larger C-terminal domain contains the active site and is the area responsible for all enzymatic activity. | + | One characteristic that makes the Variola virus so deadly is that it doesn’t require the host cell’s genetic machinery to begin replication. The reason for this is that the Variola virus codes for all the enzymes needed for its proliferation. One enzyme in particular, the type IB topoisomerase is responsible for unwinding the packaged, supercoiled, viral DNA to initiate viral replication (PENN Medicine, 2006). This is accomplished through a nick-joining mechanism of the double stranded DNA, in which a tyrosine nucleophile attacks a phosphodiester bond to form a 3’ phosphotyrosine linkage and releases a free 5’ hydroxyl group. This creates a nick to unwind supercoiling, which is then re-ligated to form relaxed DNA (Perry et al, 2010). Most mechanisms involving DNA including replication, transcription, and repair induce a certain degree of positive supercoiling. Tension due to the positive supercoiling buildup can be relieved by the enzymes topoisomerase. The variola topoisomerase 1B is an unusual topoisomerase. It is by far the smallest topoisomerase (33Kda) and only acts at specific sites that contain the sequence 5’-(T/C)CCTT-3’ in the variola genome. The topoisomerase 1B is made up of two domains that wrap around this recognition sequence forming a clamp around the DNA (Minkah et al, 2007). The smaller amino terminal domain is responsible for interactions with the DNA sequence, whereas the larger C-terminal domain contains the active site and is the area responsible for all enzymatic activity. |
[[Image:Comparison_of_Euk_and_Viral_Topo.jpg ]] | [[Image:Comparison_of_Euk_and_Viral_Topo.jpg ]] | ||
| Line 14: | Line 14: | ||
[[Image:Active_Sites_of_topo.jpg]] | [[Image:Active_Sites_of_topo.jpg]] | ||
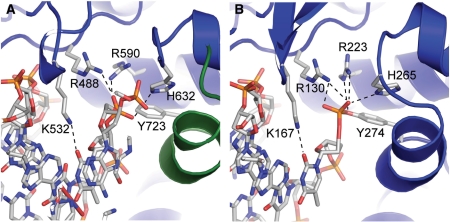
| - | In the figure above, [A] shows the amino acid residues located within the active site of eukaryotic topoisomerase 1B. [B] shows the amino acid residues located within the active site of viral topoisomerase 1B. Both active sites contain similar residues and are highly conserved among the different species. Researchers have proven that an asparagine residue can replace the histidine residue in the active site of both eukaryotic and viral topoisomerase, and the enzyme will still undergo the same cleavage mechanism. This suggests that the same cleavage and reliagation mechanisms are the same in all topoisomerase 1B’s.Type IB topoisomerase is a key target for research against the spread of smallpox because it is integral for the viruses replication process. The replication of smallpox is complicated since it doesn’t hijack the host’s genetic machinery to reproduce, this makes the disease highly virulent, and hard to specifically target for elimination by antiviral drugs. | + | In the figure above, [A] shows the amino acid residues located within the active site of eukaryotic topoisomerase 1B. [B] shows the amino acid residues located within the active site of viral topoisomerase 1B. Both active sites contain similar residues and are highly conserved among the different species. Researchers have proven that an asparagine residue can replace the histidine residue in the active site of both eukaryotic and viral topoisomerase, and the enzyme will still undergo the same cleavage mechanism. This suggests that the same cleavage and reliagation mechanisms are the same in all topoisomerase 1B’s (Baker et al, 2009).Type IB topoisomerase is a key target for research against the spread of smallpox because it is integral for the viruses replication process. The replication of smallpox is complicated since it doesn’t hijack the host’s genetic machinery to reproduce, this makes the disease highly virulent, and hard to specifically target for elimination by antiviral drugs (Berwald, 2004). |
== Relevance == | == Relevance == | ||
| Line 22: | Line 22: | ||
== Structural highlights == | == Structural highlights == | ||
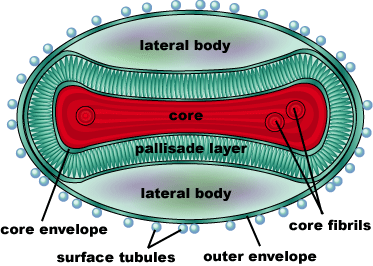
| - | Smallpox is caused by the Variola virus, a member of the genus Orthopoxvirus. It’s genome consists of a single, linear double-stranded DNA molecule made up of 186,102 base pairs. There are 10 enzymes that regulate gene expression, as well as about 100 nucleoproteins involved in transcription of the DNA. The DNA and nucleoproteins are held within a biconcave (dumbbell-shaped) core with two lateral bodies on either side. The function of the lateral bodies is unknown. The outer surface of the variola virus consists of lipids and proteins that surround this core. The variola virus consists of 90% protein, 5% lipid, and 3% DNA. | + | Smallpox is caused by the Variola virus, a member of the genus Orthopoxvirus (Smallpox, CDC). It’s genome consists of a single, linear double-stranded DNA molecule made up of 186,102 base pairs. There are 10 enzymes that regulate gene expression, as well as about 100 nucleoproteins involved in transcription of the DNA. The DNA and nucleoproteins are held within a biconcave (dumbbell-shaped) core with two lateral bodies on either side. The function of the lateral bodies is unknown. The outer surface of the variola virus consists of lipids and proteins that surround this core. The variola virus consists of 90% protein, 5% lipid, and 3% DNA (Shubhash and Parija, 2009). |
This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
Revision as of 16:05, 16 November 2015
Smallpox (Variola Virus) - Topoisomerase 1B
| |||||||||||
References
Berwald, Juli. "Variola Virus." Encyclopedia of Espionage, Intelligence, and Security. 2004.Encyclopedia.com. 28 Oct. 2015 <http://www.encyclopedia.com>.
"PENN Medicine News: Penn Researchers Determine Structure of Smallpox Virus Protein Bound to DNA." PENN Medicine News: Penn Researchers Determine Structure of Smallpox Virus Protein Bound to DNA. PENN Medicine, 4 Aug. 2006. Web. 28 Oct. 2015. <http://www.uphs.upenn.edu/news/News_Releases/aug06/smlpxenz.htm>.
Perry, Kay, Young Hwang, Frederic D. Bushman, and Gregory D. Van Duyne. "Insights from the Structure of a Smallpox Virus Topoisomerase-DNA Transition State Mimic." Structure (London, England : 1993). U.S. National Library of Medicine, n.d. Web. 28 Oct. 2015. <http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2822398/>.
“Poxvirus.” Encyclopaedia Britannica. Britannica Academic. Encyclopædia Britannica Inc., 2015. Web. 28 Oct. 2015. <http://academic.eb.com/EBchecked/topic/473442/poxvirus>.
Shubhash, and Parija. "Poxviruses." Textbook of Microbiology and Immunity. Ed. Chandra. India: Elsevior, 2009. 484. Print.
“Smallpox.” Center for Disease Control and Prevention. CDC, n.d. Web. 28 Oct. 2015. <http://www.bt.cdc.gov/agent/smallpox/index.asp>.