Caffeine
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
This is a default text for your page '''Caffeine'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | This is a default text for your page '''Caffeine'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
| - | |||
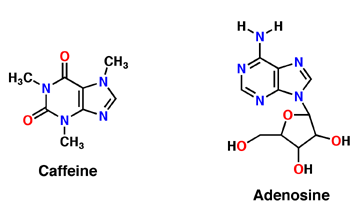
[[Image:Caffeine_and_adenosine.gif]] | [[Image:Caffeine_and_adenosine.gif]] | ||
| + | |||
== Adenosine == | == Adenosine == | ||
| Line 21: | Line 21: | ||
<Structure load='4UHR' size='350' frame='true' align='right' caption='Agonist Bound to A2A Receptor' scene='Insert optional scene name here' /> | <Structure load='4UHR' size='350' frame='true' align='right' caption='Agonist Bound to A2A Receptor' scene='Insert optional scene name here' /> | ||
| + | |||
| + | == Mechanism of Caffeine Synthesis == | ||
| + | |||
| + | Caffeine is a naturally occurring methylxanthine, purine alkaloid, synthesized by eudicot plants such as coffee, cacao, and tea (Denoeud et. al, 2014). In order to synthesize caffeine, xanthosine must undergo 3 methylation steps with the help of three NMT enzymes; xanthosine methyltransferase (XMT), theobromine synthase (MXMT), and caffeine synthase (DXMT) (Denoeud et. al, 2014). The first step of caffeine biosynthesis involves XMT converting S-adenosylmethionine (SAM) to S-adenosylhomocysteine (SAH) which removes a methyl group and adds it to the 7’-Nitrogen. This produces the intermediate 7-methyl-xanthosine which may undergo resonance to become 7-methyl-xanthine (Denoeud et. al, 2014). The second enzyme, MXMT, converts another SAM to SAH, subsequently add a methyl group to the 3’- Nitrogen on 7-methyl-xanthine. This produces theobromine which may undergo another methylation step with the help of the enzyme DXMT. DXMT converts a third SAM to SAH, adding a methyl group to the 1’-Nitrogen, yielding a caffeine molecule (Denoeud et. al, 2014). | ||
| + | |||
| + | == How Trimethylxanthine Binds to the A2A Receptor == | ||
| + | |||
| + | A2A is a transmembrane G protein in humans. Trimethylxanthine has a highly water soluble and thus when present in the system, interacts with the A2A receptor. In order for Trimethylxantine to bind to the receptor, the third and seventh transmembrane helical domains need to recognize the ligand. Trimethylxanthine can then bind. Trimethylxanthine can bind with very little discomfort, due to its similar structure, as well as its purine alkaloid structure, to adenosine. This binding will change the shape and not initiate the cascade of downstream effects that adenosine does, like opening of ion channels and slowing of activity. Concentrate of free adenosine increases extracellularly, when trimethylxanthine is bound. The cAMP increases when adenosine is bound and (). ERK1 and ERK2 are kinases, composed of serine and threonine, of the GMGC group that regulation of cell growth and differentiation, and if adenosine was bound, this cascade of events would occur, but when Trimethylxanthine is bound, this regulation does not occur. | ||
| + | |||
| + | == See Also == | ||
| + | |||
| + | http://www.nature.com/nrc/journal/v13/n12/fig_tab/nrc3613_F1.html | ||
| + | |||
| + | http://www.rcsb.org/pdb/explore/explore.do?structureId=4UHR | ||
| + | |||
| + | http://www.rcsb.org/pdb/explore/explore.do?structureId=3RFM | ||
| + | |||
| + | http://www.edb.utexas.edu/ssn/SN%20PDF/Caffeine-Exercise%20Perform.PDF | ||
| + | |||
| + | http://www.ncbi.nlm.nih.gov/pubmed/20164566 | ||
| + | |||
| + | http://www.pnas.org/content/112/25/7833.short | ||
| + | |||
| + | http://www.ncbi.nlm.nih.gov/pubmed/1356551 | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 02:34, 17 November 2015
Caffeine
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
Proteopedia Page Contributors and Editors (what is this?)
Mackenzie Kincer, Sidiqullah Halim, Alexander Berchansky, Michal Harel