We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1172
From Proteopedia
(Difference between revisions)
| Line 33: | Line 33: | ||
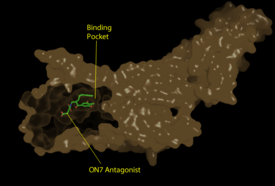
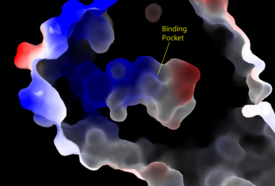
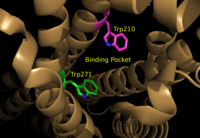
A model for lipid agonist binding generated through molecular modeling was used to dock two of the cannabinoid receptor CB<sub>1</sub>'s most abundant endogenous ligands into the LPA<sub>1</sub> binding pocket. <ref name="regpeps">PMID: 26091040</ref> Rotameric shifts of Trp210 and Trp271 leading to expansion of the binding pocket and the exposure of the π clouds of their indole rings provided favorable interactions with the double bonds of the phosphorylated cannabinoid ligands. This favorable binding provides evidence that the hydrophobic binding pockets of LPA<sub>1</sub> and CB<sub>1</sub> are able to favorably bind the same poly-unsaturated acyl chains with metabolically interconvertible head groups. <ref name="regpeps">PMID: 26091040</ref> | A model for lipid agonist binding generated through molecular modeling was used to dock two of the cannabinoid receptor CB<sub>1</sub>'s most abundant endogenous ligands into the LPA<sub>1</sub> binding pocket. <ref name="regpeps">PMID: 26091040</ref> Rotameric shifts of Trp210 and Trp271 leading to expansion of the binding pocket and the exposure of the π clouds of their indole rings provided favorable interactions with the double bonds of the phosphorylated cannabinoid ligands. This favorable binding provides evidence that the hydrophobic binding pockets of LPA<sub>1</sub> and CB<sub>1</sub> are able to favorably bind the same poly-unsaturated acyl chains with metabolically interconvertible head groups. <ref name="regpeps">PMID: 26091040</ref> | ||
| - | [[Image:TrpRotamericShiftsCM.png|200 px|left|thumb|Figure 1: | + | [[Image:TrpRotamericShiftsCM.png|200 px|left|thumb|Figure 1: Illustration of key LPA<sub>1</sub> binding pocket residues Trp210 and Trp271. These residues provide rotameric shifts and expansion that allow for favorable interactions with phosphorylated cannabinoid ligands.]] |
== Disease Relevance == | == Disease Relevance == | ||
Revision as of 22:03, 18 April 2016
| This Sandbox is Reserved from Jan 11 through August 12, 2016 for use in the course CH462 Central Metabolism taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1160 through Sandbox Reserved 1184. |
To get started:
More help: Help:Editing |
Lysophosphatidic Acid Receptor 1
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 Chrencik JE, Roth CB, Terakado M, Kurata H, Omi R, Kihara Y, Warshaviak D, Nakade S, Asmar-Rovira G, Mileni M, Mizuno H, Griffith MT, Rodgers C, Han GW, Velasquez J, Chun J, Stevens RC, Hanson MA. Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1. Cell. 2015 Jun 18;161(7):1633-43. doi: 10.1016/j.cell.2015.06.002. PMID:26091040 doi:http://dx.doi.org/10.1016/j.cell.2015.06.002
- ↑ 2.0 2.1 Yung, Y. C., N. C. Stoddard, and J. Chun. "LPA Receptor Signaling: Pharmacology, Physiology, and Pathophysiology." The Journal of Lipid Research 55.7 (2014): 1192-214. Web. 17 Feb. 2016.'
- ↑ 3.0 3.1 Chun, J., Hla, T., Spiegel, S., and Moolenaar, W.H. “Lysophospholipid Receptors: Signaling and Biochemistry.” John Wiley & Sons, Inc. (2013) pp.i-xviii. 5 Feb. 2016.'
- ↑ Hernández-Méndez, Aurelio, Rocío Alcántara-Hernández, and J. Adolfo García-Sáinz. "Lysophosphatidic Acid LPA1-3 Receptors: Signaling, Regulation and in Silico Analysis of Their Putative Phosphorylation Sites." Receptors & Clinical Investigation Receptor Clin Invest 1.3 (2014). Web. 15 Feb. 2016.'
- ↑ Anliker B, Choi JW, Lin ME, Gardell SE, Rivera RR, Kennedy G, Chun J. Lysophosphatidic acid (LPA) and its receptor, LPA1 , influence embryonic schwann cell migration, myelination, and cell-to-axon segregation. Glia. 2013 Dec;61(12):2009-22. doi: 10.1002/glia.22572. Epub 2013 Sep 24. PMID:24115248 doi:http://dx.doi.org/10.1002/glia.22572
- ↑ Chun, E., Thompson, A.A., Lui, W., Roth, C.B., Griffith, M.T., Katritch, V., Kunken, J., Xu, F., Cherezov, V., Hanson, M.A., and Stevens, R.C. “Fusion partner tool chest for the stabilization and crystallization of G protein-coupled receptors.” Structure 20, (2012) 967-976.'
- ↑ Van Durme, J., Horn, F., Costagliola, S., Vriend, G., and Vassart, G. “GRIS: glycoprotein-hormone receptor information system.” Mol. (2006) Endocrinol. 20, 2247-2255'
- ↑ 8.0 8.1 8.2 8.3 Lin ME, Herr DR, Chun J. Lysophosphatidic acid (LPA) receptors: signaling properties and disease relevance. Prostaglandins Other Lipid Mediat. 2010 Apr;91(3-4):130-8. doi:, 10.1016/j.prostaglandins.2009.02.002. Epub 2009 Mar 4. PMID:20331961 doi:http://dx.doi.org/10.1016/j.prostaglandins.2009.02.002
- ↑ 9.0 9.1 9.2 Justus CR, Dong L, Yang LV. Acidic tumor microenvironment and pH-sensing G protein-coupled receptors. Front Physiol. 2013 Dec 5;4:354. doi: 10.3389/fphys.2013.00354. PMID:24367336 doi:http://dx.doi.org/10.3389/fphys.2013.00354