We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1172
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
== Structure == | == Structure == | ||
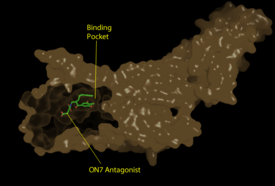
[[Image:Pic2proteo3.png|275 px|left|thumb|Figure 1: Surface representation of the LPA<sub>1</sub> receptor in tan interacting with its antagonist, ON7, shown in green and red sticks.The exterior of the protein was partially cut away to display the interior binding pocket.]] | [[Image:Pic2proteo3.png|275 px|left|thumb|Figure 1: Surface representation of the LPA<sub>1</sub> receptor in tan interacting with its antagonist, ON7, shown in green and red sticks.The exterior of the protein was partially cut away to display the interior binding pocket.]] | ||
| - | The LPA<sub>1</sub> receptor protein is composed of 364 amino acids with a molecular weight of approximately 41 kDa. Common to all G-protein coupled receptors, LPA<sub>1</sub> contains seven [http://kinemage.biochem.duke.edu/teaching/anatax/html/anatax.2a.html alpha helices] which make up the seven transmembrane spanning domains with three intracellular loops and three extracellular loops.<ref name = 'Hernández-Méndez'>Hernández-Méndez, Aurelio, Rocío Alcántara-Hernández, and J. Adolfo García-Sáinz. "Lysophosphatidic Acid LPA1-3 Receptors: Signaling, Regulation and in Silico Analysis of Their Putative Phosphorylation Sites." Receptors & Clinical Investigation Receptor Clin Invest 1.3 (2014). Web. 15 Feb. 2016.' </ref> | + | The LPA<sub>1</sub> receptor protein is composed of 364 amino acids with a molecular weight of approximately 41 kDa. Common to all G-protein coupled receptors, LPA<sub>1</sub> contains seven [http://kinemage.biochem.duke.edu/teaching/anatax/html/anatax.2a.html alpha helices] which make up the seven transmembrane spanning domains with three intracellular loops and three extracellular loops.<ref name = 'Hernández-Méndez'>Hernández-Méndez, Aurelio, Rocío Alcántara-Hernández, and J. Adolfo García-Sáinz. "Lysophosphatidic Acid LPA1-3 Receptors: Signaling, Regulation and in Silico Analysis of Their Putative Phosphorylation Sites." Receptors & Clinical Investigation Receptor Clin Invest 1.3 (2014). Web. 15 Feb. 2016.' </ref> Within these helices is a binding pocket which stabilizes the binding of its ligand, LPA (Figure 1). |
=== Key Ligand Interactions === | === Key Ligand Interactions === | ||
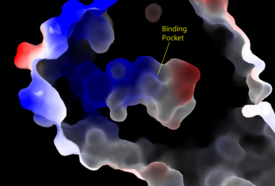
[[Image:Amphbindingfinal.png|275 px|right|thumb|Figure 2: Electrostatic illustration of the amphipathic binding pocket of the LPA<sub>1</sub> receptor. This binding pocket was revealed by cutting away the exterior or the protein. This binding pocket, located in the interior of the protein, has both polar and nonpolar regions. The blue and red coloration highlight the positively and negatively charged regions, respectively, and the white color shows the nonpolar region of the binding pocket.]] | [[Image:Amphbindingfinal.png|275 px|right|thumb|Figure 2: Electrostatic illustration of the amphipathic binding pocket of the LPA<sub>1</sub> receptor. This binding pocket was revealed by cutting away the exterior or the protein. This binding pocket, located in the interior of the protein, has both polar and nonpolar regions. The blue and red coloration highlight the positively and negatively charged regions, respectively, and the white color shows the nonpolar region of the binding pocket.]] | ||
| Line 15: | Line 15: | ||
== Function == | == Function == | ||
| - | The LPA<sub>1</sub> receptor is present in nearly all cells and tissues, and deletion of the LPA<sub>1</sub> receptor has physiological effects on every organ system, indicating its wide range of functions. Specifically, this receptor initiates downstream signaling cascades with three G<sub>α</sub> proteins: G<sub>i/o</sub>, G<sub>q/11</sub>, and G<sub>12/13</sub>. Specifically, G<sub>α</sub> proteins begin signaling cascades that activate [https://en.wikipedia.org/wiki/Phospholipase_C phospholipase C] and [[MAPK]]s that signal for cell proliferation, survival, and migration. <ref name = 'Yung'>Yung, Y. C., N. C. Stoddard, and J. Chun. "LPA Receptor Signaling: Pharmacology, Physiology, and Pathophysiology." The Journal of Lipid Research 55.7 (2014): 1192-214. Web. 17 Feb. 2016.' </ref> | + | The LPA<sub>1</sub> receptor is present in nearly all cells and tissues, and deletion of the LPA<sub>1</sub> receptor has physiological effects on every organ system, indicating its wide range of functions. Specifically, this receptor initiates downstream signaling cascades with three G<sub>α</sub> proteins: G<sub>i/o</sub>, G<sub>q/11</sub>, and G<sub>12/13</sub>. Specifically, G<sub>α</sub> proteins begin signaling cascades that activate [https://en.wikipedia.org/wiki/Phospholipase_C phospholipase C] and [[MAPK]]s that signal for cell proliferation, survival, and migration. <ref name = 'Yung'>Yung, Y. C., N. C. Stoddard, and J. Chun. "LPA Receptor Signaling: Pharmacology, Physiology, and Pathophysiology." The Journal of Lipid Research 55.7 (2014): 1192-214. Web. 17 Feb. 2016.' </ref> Despite this receptor being expressed ubiquitously, LPA<sub>1</sub> is highly expressed in neural tissue, aiding in several different neurodevelopmental processes including growth and folding of the [https://www.dartmouth.edu/~rswenson/NeuroSci/chapter_11.html cerebral cortex] and the growth, survival, and migration of neural [https://en.wikipedia.org/wiki/Progenitor_cell progenitor cells].<ref name = 'Chun'>Chun, J., Hla, T., Spiegel, S., and Moolenaar, W.H. “Lysophospholipid Receptors: Signaling and Biochemistry.” John Wiley & Sons, Inc. (2013) pp.i-xviii. 5 Feb. 2016.' </ref> Finally, LPA<sub>1</sub> receptors expressed in neural tissue act through a signaling pathway with the [https://en.wikipedia.org/wiki/RAC1 Rac1] G-protein to aid in Schwann cell migration and myelination.<ref name="number5">PMID: 24115248</ref> |
| Line 22: | Line 22: | ||
=== Sphingosine 1-Phosphate Receptor === | === Sphingosine 1-Phosphate Receptor === | ||
| - | LPA<sub>1</sub> belongs to the EDG (endothelial differentiation gene) family of [https://en.wikipedia.org/wiki/Lysophospholipid_receptor lysophospholipid receptors]. This family also includes the [https://en.wikipedia.org/wiki/S1PR1 sphingosine 1-phosphate receptor 1] (S1P<sub>1</sub>), which | + | LPA<sub>1</sub> belongs to the EDG (endothelial differentiation gene) family of [https://en.wikipedia.org/wiki/Lysophospholipid_receptor lysophospholipid receptors]. This family also includes the [https://en.wikipedia.org/wiki/S1PR1 sphingosine 1-phosphate receptor 1] (S1P<sub>1</sub>), which shares many structural similarities to LPA<sub>1</sub>. In fact, the transmembrane regions share a sequence identity of 41%. <ref name = 'Chun, E.'>Chun, E., Thompson, A.A., Lui, W., Roth, C.B., Griffith, M.T., Katritch, V., Kunken, J., Xu, F., Cherezov, V., Hanson, M.A., and Stevens, R.C. “Fusion partner tool chest for the stabilization and crystallization of G protein-coupled receptors.” Structure 20, (2012) 967-976.' </ref> A defining difference between these two receptors is their mode of ligand access to the binding site. The hydrophobic [https://en.wikipedia.org/wiki/Sphingosine-1-phosphate S1P ligand] enters S1P<sub>1</sub> via the membrane. LPA<sub>1</sub> differs and utilizes an extracellular opening that allows LPA access from the extracellular space (Figure 2). <ref name="regpeps">PMID: 26091040</ref> Structural evidence for this altered ligand binding pathway includes global changes in the positioning of the extracellular loops (ECL) and transmembrane helices (TM). Specifically, a slight divergence of <scene name='72/721543/Tmvii_and_tmi/1'>TMI</scene>, which is positioned 3 Å closer to TMVII compared to S1P<sub>1</sub>, and a repositioning of <scene name='72/721543/Ecl_regions/1'>ECL3</scene>, resulting in a divergence of 8 Å from S1P<sub>1</sub> result in ligand access via the extracellular space. <ref name="regpeps">PMID: 26091040</ref> This narrowing of the gap between TMI and TMVII blocks membrane ligand access, while the greater distance between ECL3 and the other extracellular loops promotes extracellular access for LPA<sub>1</sub>. Additionally, ECL0 is helical in S1P<sub>1</sub>, but <scene name='72/721543/Ecl02ndstructure/1'>lacks secondary structure</scene> in LPA<sub>1</sub>. This increased flexibility that results further promotes favorable access from the extracellular space. <ref name="regpeps">PMID: 26091040</ref> |
| Line 28: | Line 28: | ||
=== Endocannabinoid Receptor 1 === | === Endocannabinoid Receptor 1 === | ||
| - | LPA<sub>1</sub> also is closely related to the first of the [http://www.nature.com/ijo/journal/v30/n1s/full/0803272a.html cannabinoid receptors]. This close relation gives CB<sub>1</sub> ([[Cannabinoid Receptor 1]]) the ability to bind to analogs of LPA and vice versa, which opens the possibility of metabolic crosstalk between the two signaling systems. This connection is made possible through ligand phosphorylation and dephosphorylation <ref name="regpeps">PMID: 26091040</ref> | + | LPA<sub>1</sub> also is closely related to the first of the [http://www.nature.com/ijo/journal/v30/n1s/full/0803272a.html cannabinoid receptors]. This close relation gives CB<sub>1</sub> ([[Cannabinoid Receptor 1]]) the ability to bind to analogs of LPA and vice versa, which opens the possibility of metabolic crosstalk between the two signaling systems. This connection is made possible through ligand phosphorylation and dephosphorylation. <ref name="regpeps">PMID: 26091040</ref> Specifically, complementary access to the LPA<sub>1</sub> binding pocket can be achieved by phosphorylated CB<sub>1</sub> ligand analogs, while complementary access to the CB<sub>1</sub> binding site requires dephosphorylation of LPA<sub>1</sub> ligand analogs. In both cases, a ligand could serve as a primary [https://en.wikipedia.org/wiki/Selective_receptor_modulator receptor modulator] and a simultaneous [https://en.wikipedia.org/wiki/Prodrug prodrug] for a different receptor. <ref name="regpeps">PMID: 26091040</ref> |
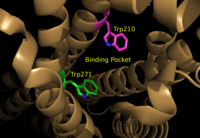
<scene name='72/721543/Asp129_and_trp210/2'>Residues Asp129 and Trp210</scene> located within the hydrophobic binding pocket of LPA<sub>1</sub> may share responsibility for the preference for long unsaturated acyl chains. These residues are also interesting in regard to GPCR phylogenic evolution. <ref name="regpeps">PMID: 26091040</ref> Trp210 specifically only occurs in this position in 1% of all class A GPCR receptors and is unique to lysophospholipid and cannabinoid receptors. <ref name = 'Van Durme'>Van Durme, J., Horn, F., Costagliola, S., Vriend, G., and Vassart, G. “GRIS: glycoprotein-hormone receptor information system.” Mol. (2006) Endocrinol. 20, 2247-2255' </ref> | <scene name='72/721543/Asp129_and_trp210/2'>Residues Asp129 and Trp210</scene> located within the hydrophobic binding pocket of LPA<sub>1</sub> may share responsibility for the preference for long unsaturated acyl chains. These residues are also interesting in regard to GPCR phylogenic evolution. <ref name="regpeps">PMID: 26091040</ref> Trp210 specifically only occurs in this position in 1% of all class A GPCR receptors and is unique to lysophospholipid and cannabinoid receptors. <ref name = 'Van Durme'>Van Durme, J., Horn, F., Costagliola, S., Vriend, G., and Vassart, G. “GRIS: glycoprotein-hormone receptor information system.” Mol. (2006) Endocrinol. 20, 2247-2255' </ref> | ||
Revision as of 22:26, 18 April 2016
| This Sandbox is Reserved from Jan 11 through August 12, 2016 for use in the course CH462 Central Metabolism taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1160 through Sandbox Reserved 1184. |
To get started:
More help: Help:Editing |
Lysophosphatidic Acid Receptor 1
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 Chrencik JE, Roth CB, Terakado M, Kurata H, Omi R, Kihara Y, Warshaviak D, Nakade S, Asmar-Rovira G, Mileni M, Mizuno H, Griffith MT, Rodgers C, Han GW, Velasquez J, Chun J, Stevens RC, Hanson MA. Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1. Cell. 2015 Jun 18;161(7):1633-43. doi: 10.1016/j.cell.2015.06.002. PMID:26091040 doi:http://dx.doi.org/10.1016/j.cell.2015.06.002
- ↑ 2.0 2.1 Yung, Y. C., N. C. Stoddard, and J. Chun. "LPA Receptor Signaling: Pharmacology, Physiology, and Pathophysiology." The Journal of Lipid Research 55.7 (2014): 1192-214. Web. 17 Feb. 2016.'
- ↑ 3.0 3.1 Chun, J., Hla, T., Spiegel, S., and Moolenaar, W.H. “Lysophospholipid Receptors: Signaling and Biochemistry.” John Wiley & Sons, Inc. (2013) pp.i-xviii. 5 Feb. 2016.'
- ↑ Hernández-Méndez, Aurelio, Rocío Alcántara-Hernández, and J. Adolfo García-Sáinz. "Lysophosphatidic Acid LPA1-3 Receptors: Signaling, Regulation and in Silico Analysis of Their Putative Phosphorylation Sites." Receptors & Clinical Investigation Receptor Clin Invest 1.3 (2014). Web. 15 Feb. 2016.'
- ↑ Anliker B, Choi JW, Lin ME, Gardell SE, Rivera RR, Kennedy G, Chun J. Lysophosphatidic acid (LPA) and its receptor, LPA1 , influence embryonic schwann cell migration, myelination, and cell-to-axon segregation. Glia. 2013 Dec;61(12):2009-22. doi: 10.1002/glia.22572. Epub 2013 Sep 24. PMID:24115248 doi:http://dx.doi.org/10.1002/glia.22572
- ↑ Chun, E., Thompson, A.A., Lui, W., Roth, C.B., Griffith, M.T., Katritch, V., Kunken, J., Xu, F., Cherezov, V., Hanson, M.A., and Stevens, R.C. “Fusion partner tool chest for the stabilization and crystallization of G protein-coupled receptors.” Structure 20, (2012) 967-976.'
- ↑ Van Durme, J., Horn, F., Costagliola, S., Vriend, G., and Vassart, G. “GRIS: glycoprotein-hormone receptor information system.” Mol. (2006) Endocrinol. 20, 2247-2255'
- ↑ 8.0 8.1 8.2 8.3 Lin ME, Herr DR, Chun J. Lysophosphatidic acid (LPA) receptors: signaling properties and disease relevance. Prostaglandins Other Lipid Mediat. 2010 Apr;91(3-4):130-8. doi:, 10.1016/j.prostaglandins.2009.02.002. Epub 2009 Mar 4. PMID:20331961 doi:http://dx.doi.org/10.1016/j.prostaglandins.2009.02.002
- ↑ 9.0 9.1 9.2 Justus CR, Dong L, Yang LV. Acidic tumor microenvironment and pH-sensing G protein-coupled receptors. Front Physiol. 2013 Dec 5;4:354. doi: 10.3389/fphys.2013.00354. PMID:24367336 doi:http://dx.doi.org/10.3389/fphys.2013.00354