Sandbox reserved 1169
From Proteopedia
(Difference between revisions)
| Line 16: | Line 16: | ||
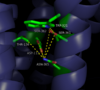
Binding of NTS to the binding site on NTSR1 is enriched by <scene name='72/721539/Binding_pocket_surface/4'>charge complementarity</scene> (PDB code:[http://www.rcsb.org/pdb/explore/explore.do?structureId=4GRV 4GRV)]between the positive NTS arginine side chains and the [https://en.wikipedia.org/wiki/Electronegativity electronegative] pocket. The protein is colored by charge: <font color='#FF0000'>negative</font> and <font color='#0000CD'>positive</font>. Two of NTS's arginine residues are colored <font color='#0000CD'>blue</font>. In addition, the C-terminus of <font color='#32CD32'>NTS</font> forms a <scene name='72/721539/Binding_site_charges/4'>salt bridge</scene> (PDB code:[http://www.rcsb.org/pdb/explore/explore.do?structureId=4GRV 4GRV)] with Arg328 of <font color='#A9A9A9'>NTSR1</font>. Three [https://en.wikipedia.org/wiki/Hydrogen_bond hydrogen bonds] are made between the side chains of NTS and the receptor while most of the interactions are a result of [https://en.wikipedia.org/wiki/Van_der_Waals_force Van der Waals] interactions. The binding pocket is partially capped by a <scene name='72/721539/B-hairpin_loop/1'>Β-hairpin loop</scene> at the proximal end of the receptor's N-terminus.<ref name="White"/> The interactions in the binding site cause a wide spread conformational change in the receptor leading to the receptor to adopt an active conformation. This active conformation allows for the activation of the associated intracellular G-protein. | Binding of NTS to the binding site on NTSR1 is enriched by <scene name='72/721539/Binding_pocket_surface/4'>charge complementarity</scene> (PDB code:[http://www.rcsb.org/pdb/explore/explore.do?structureId=4GRV 4GRV)]between the positive NTS arginine side chains and the [https://en.wikipedia.org/wiki/Electronegativity electronegative] pocket. The protein is colored by charge: <font color='#FF0000'>negative</font> and <font color='#0000CD'>positive</font>. Two of NTS's arginine residues are colored <font color='#0000CD'>blue</font>. In addition, the C-terminus of <font color='#32CD32'>NTS</font> forms a <scene name='72/721539/Binding_site_charges/4'>salt bridge</scene> (PDB code:[http://www.rcsb.org/pdb/explore/explore.do?structureId=4GRV 4GRV)] with Arg328 of <font color='#A9A9A9'>NTSR1</font>. Three [https://en.wikipedia.org/wiki/Hydrogen_bond hydrogen bonds] are made between the side chains of NTS and the receptor while most of the interactions are a result of [https://en.wikipedia.org/wiki/Van_der_Waals_force Van der Waals] interactions. The binding pocket is partially capped by a <scene name='72/721539/B-hairpin_loop/1'>Β-hairpin loop</scene> at the proximal end of the receptor's N-terminus.<ref name="White"/> The interactions in the binding site cause a wide spread conformational change in the receptor leading to the receptor to adopt an active conformation. This active conformation allows for the activation of the associated intracellular G-protein. | ||
=== Hydrophobic Stacking === | === Hydrophobic Stacking === | ||
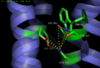
| - | A major player in the transduction of the extracellular signal to the intracellular G protein is the <scene name='72/727765/Hydrogen_bonding_network/4'>hydrogen bonding network</scene> (PDB code:[http://www.rcsb.org/pdb/explore/explore.do?structureId=4GRV 4GRV)]that links the bound <font color='#32CD32'>hormone</font> with the [https://en.wikipedia.org/wiki/Hydrophobe hydrophobic] core of the <font color='#A9A9A9'>neurotensin receptor</font>. The carboxylate of Leu13 of NTS forms a hydrogen bond network with R327, R328, and Y324. The Tyr324, in turn, is brought into an orientation to make the formation of a <scene name='72/727765/Hydrophobic_stacking_4xee/2'>hydrophobic stacking</scene> (PDB Code:[http://www.rcsb.org/pdb/explore/explore.do?structureId=4XEE 4XEE]) network between F358, W321, A157, and F317 possible.<ref name="Krumm">PMID:23051748</ref> The effects that this network has on the activation of the intracellular G-protein was examined by the [https://en.wikipedia.org/wiki/Mutagenesis mutagenesis] of amino acids involved in this network. Mutagenesis of <scene name='72/727765/Overall_structure/12'>A86L</scene>, <scene name='72/727765/Overall_structure/13'>E166A</scene>, <scene name='72/727765/Overall_structure/14'>G125L</scene>, <scene name='72/727765/Overall_structure/15'>L310A</scene>, <scene name='72/727765/Overall_structure/16'>F358A</scene>, and <scene name='72/727765/Overall_structure/17'>V360A</scene> (PDB code:[http://www.rcsb.org/pdb/explore/explore.do?structureId=4GRV 4GRV)] showed that when this interaction was disrupted, the receptor no longer was able to activate the G-protein. <ref name="Krumm"/> This discovery lead to the conclusion that the conformational changes caused by this stacking allows for the signal to be moved from the extracellular binding site through the transmembrane helices of the receptor to the intracellular region activating the G-protein. | + | A major player in the transduction of the extracellular signal to the intracellular G-protein is the <scene name='72/727765/Hydrogen_bonding_network/4'>hydrogen bonding network</scene> (PDB code:[http://www.rcsb.org/pdb/explore/explore.do?structureId=4GRV 4GRV)]that links the bound <font color='#32CD32'>hormone</font> with the [https://en.wikipedia.org/wiki/Hydrophobe hydrophobic] core of the <font color='#A9A9A9'>neurotensin receptor</font>. The carboxylate of Leu13 of NTS forms a hydrogen bond network with R327, R328, and Y324. The Tyr324, in turn, is brought into an orientation to make the formation of a <scene name='72/727765/Hydrophobic_stacking_4xee/2'>hydrophobic stacking</scene> (PDB Code:[http://www.rcsb.org/pdb/explore/explore.do?structureId=4XEE 4XEE]) network between F358, W321, A157, and F317 possible.<ref name="Krumm">PMID:23051748</ref> The effects that this network has on the activation of the intracellular G-protein was examined by the [https://en.wikipedia.org/wiki/Mutagenesis mutagenesis] of amino acids involved in this network. Mutagenesis of <scene name='72/727765/Overall_structure/12'>A86L</scene>, <scene name='72/727765/Overall_structure/13'>E166A</scene>, <scene name='72/727765/Overall_structure/14'>G125L</scene>, <scene name='72/727765/Overall_structure/15'>L310A</scene>, <scene name='72/727765/Overall_structure/16'>F358A</scene>, and <scene name='72/727765/Overall_structure/17'>V360A</scene> (PDB code:[http://www.rcsb.org/pdb/explore/explore.do?structureId=4GRV 4GRV)] showed that when this interaction was disrupted, the receptor no longer was able to activate the G-protein. <ref name="Krumm"/> This discovery lead to the conclusion that the conformational changes caused by this stacking allows for the signal to be moved from the extracellular binding site through the transmembrane helices of the receptor to the intracellular region activating the G-protein. |
== Sodium Binding Pocket == | == Sodium Binding Pocket == | ||
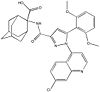
[[Image:4XEE closed sodium pocket.png|100 px|left|thumb|Figure 2: Closed form of sodium binding pocket that caps the entrance of sodium into the top of the binding pocket. (PDB Code:[http://www.rcsb.org/pdb/explore/explore.do?structureId=4XEE 4XEE])]] | [[Image:4XEE closed sodium pocket.png|100 px|left|thumb|Figure 2: Closed form of sodium binding pocket that caps the entrance of sodium into the top of the binding pocket. (PDB Code:[http://www.rcsb.org/pdb/explore/explore.do?structureId=4XEE 4XEE])]] | ||
Revision as of 07:04, 19 April 2016
Neurotensin Receptor (NTSR1)
References
- ↑ Millar RP, Newton CL. The year in G protein-coupled receptor research. Mol Endocrinol. 2010 Jan;24(1):261-74. Epub 2009 Dec 17. PMID:20019124 doi:10.1210/me.2009-0473
- ↑ Gui X, Carraway RE. Enhancement of jejunal absorption of conjugated bile acid by neurotensin in rats. Gastroenterology. 2001 Jan;120(1):151-60. PMID:11208724
- ↑ Selivonenko VG. [The interrelationship between electrolytes and phase analysis of systole in toxic goiter]. Probl Endokrinol (Mosk). 1975 Jan-Feb;21(1):19-23. PMID:1173461
- ↑ Fang Y, Lahiri J, Picard L. G protein-coupled receptor microarrays for drug discovery. Drug Discov Today. 2004 Dec 15;9(24 Suppl):S61-7. PMID:23573662
- ↑ 5.0 5.1 White JF, Noinaj N, Shibata Y, Love J, Kloss B, Xu F, Gvozdenovic-Jeremic J, Shah P, Shiloach J, Tate CG, Grisshammer R. Structure of the agonist-bound neurotensin receptor. Nature. 2012 Oct 25;490(7421):508-13. doi: 10.1038/nature11558. Epub 2012 Oct 10. PMID:23051748 doi:http://dx.doi.org/10.1038/nature11558
- ↑ Carraway R, Leeman SE. The isolation of a new hypotensive peptide, neurotensin, from bovine hypothalami. J Biol Chem. 1973 Oct 10;248(19):6854-61. PMID:4745447
- ↑ Kitabgi P. Neurotensin modulates dopamine neurotransmission at several levels along brain dopaminergic pathways. Neurochem Int. 1989;14(2):111-9. PMID:20504406
- ↑ Mustain WC, Rychahou PG, Evers BM. The role of neurotensin in physiologic and pathologic processes. Curr Opin Endocrinol Diabetes Obes. 2011 Feb;18(1):75-82. doi:, 10.1097/MED.0b013e3283419052. PMID:21124211 doi:http://dx.doi.org/10.1097/MED.0b013e3283419052
- ↑ Vincent JP, Mazella J, Kitabgi P. Neurotensin and neurotensin receptors. Trends Pharmacol Sci. 1999 Jul;20(7):302-9. PMID:10390649
- ↑ 10.0 10.1 10.2 White JF, Noinaj N, Shibata Y, Love J, Kloss B, Xu F, Gvozdenovic-Jeremic J, Shah P, Shiloach J, Tate CG, Grisshammer R. Structure of the agonist-bound neurotensin receptor. Nature. 2012 Oct 25;490(7421):508-13. doi: 10.1038/nature11558. Epub 2012 Oct 10. PMID:23051748 doi:http://dx.doi.org/10.1038/nature11558
- ↑ Katritch V, Fenalti G, Abola EE, Roth BL, Cherezov V, Stevens RC. Allosteric sodium in class A GPCR signaling. Trends Biochem Sci. 2014 May;39(5):233-44. doi: 10.1016/j.tibs.2014.03.002. Epub , 2014 Apr 21. PMID:24767681 doi:http://dx.doi.org/10.1016/j.tibs.2014.03.002
- ↑ 12.0 12.1 Valerie NC, Casarez EV, Dasilva JO, Dunlap-Brown ME, Parsons SJ, Amorino GP, Dziegielewski J. Inhibition of neurotensin receptor 1 selectively sensitizes prostate cancer to ionizing radiation. Cancer Res. 2011 Nov 1;71(21):6817-26. doi: 10.1158/0008-5472.CAN-11-1646. Epub, 2011 Sep 8. PMID:21903767 doi:http://dx.doi.org/10.1158/0008-5472.CAN-11-1646
- ↑ Kisfalvi K, Eibl G, Sinnett-Smith J, Rozengurt E. Metformin disrupts crosstalk between G protein-coupled receptor and insulin receptor signaling systems and inhibits pancreatic cancer growth. Cancer Res. 2009 Aug 15;69(16):6539-45. doi: 10.1158/0008-5472.CAN-09-0418. PMID:19679549 doi:http://dx.doi.org/10.1158/0008-5472.CAN-09-0418