Gluten
From Proteopedia
(Difference between revisions)
| Line 37: | Line 37: | ||
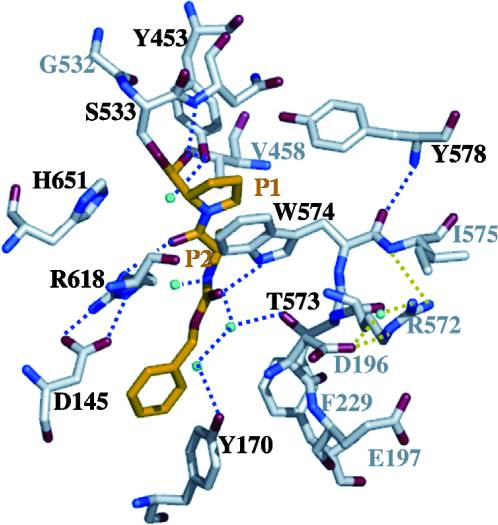
[http://en.wikipedia.org/wiki/Prolyl_endopeptidase Prolyl endopeptidases] (PEPs) are a family of [http://en.wikipedia.org/wiki/Serine_protease Serine proteases] that help accelerate the breakdown of peptides by cleaving after proline residues. Since gluten is a proline-rich complex, these enzymes are being explored as a therapy to help treat individuals with celiac disease, since these individuals aren’t able to break down gluten naturally. <ref name="shan">Shan, L., I. I. Mathews, and C. Khosla. "Structural and Mechanistic Analysis of Two Prolyl nEndopeptidases: Role of Interdomain Dynamics in Catalysis and Specificity." Proceedings of the National Academy of Sciences 102.10 (2005): 3599-604. Web.</ref> | [http://en.wikipedia.org/wiki/Prolyl_endopeptidase Prolyl endopeptidases] (PEPs) are a family of [http://en.wikipedia.org/wiki/Serine_protease Serine proteases] that help accelerate the breakdown of peptides by cleaving after proline residues. Since gluten is a proline-rich complex, these enzymes are being explored as a therapy to help treat individuals with celiac disease, since these individuals aren’t able to break down gluten naturally. <ref name="shan">Shan, L., I. I. Mathews, and C. Khosla. "Structural and Mechanistic Analysis of Two Prolyl nEndopeptidases: Role of Interdomain Dynamics in Catalysis and Specificity." Proceedings of the National Academy of Sciences 102.10 (2005): 3599-604. Web.</ref> | ||
| - | < | + | <table align="left"><tr><td> |
| + | [[Image:Proteopedia article image 2.jpg]] | ||
| + | </td></tr><tr><td> | ||
| + | Copyright 2005, National Academy of Sciences<ref name="pnas">"Permission is not required to use original figures or tables for noncommercial and educational use",[https://www.pnas.org/page/about/rights-permissions Rights & Permissions], Proceedings of the National Aademy of Sciences, USA.</ref>. | ||
| + | </td></tr></table> | ||
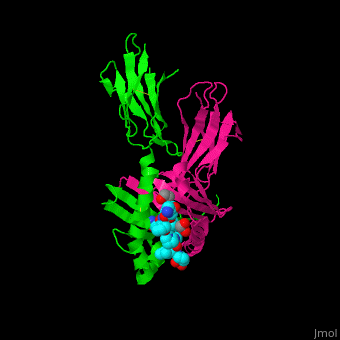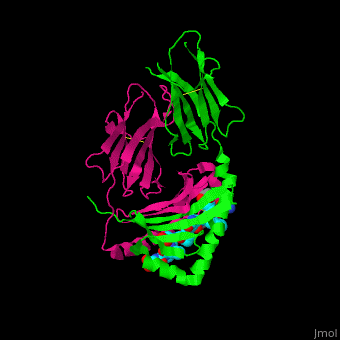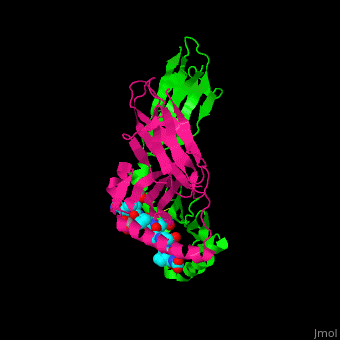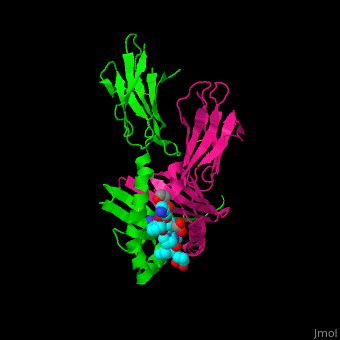
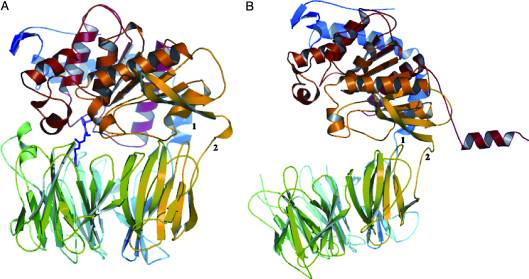
A study was done to explore the structural features of two bacterial PEPs with the purpose of evaluating their ability to cleave proline-rich residues. One was isolated from [https://en.wikipedia.org/wiki/Myxococcus_xanthus ''Myxococcus xanthus'']with a <scene name='71/719201/Bound_pep/1'> bound enzyme inhibitor </scene> and one was isolated from [https://en.wikipedia.org/wiki/Sphingomonas ''Sphingomonas capsulata''] in an <scene name='71/719201/Unbound_pep/1'> unbound state </scene>. Both PEPs have two domains: a catalytic binding site that forms interactions with the substrate and the propeller domain that acts as a sort of stabilizing clamp structure. <ref name="shan" /> | A study was done to explore the structural features of two bacterial PEPs with the purpose of evaluating their ability to cleave proline-rich residues. One was isolated from [https://en.wikipedia.org/wiki/Myxococcus_xanthus ''Myxococcus xanthus'']with a <scene name='71/719201/Bound_pep/1'> bound enzyme inhibitor </scene> and one was isolated from [https://en.wikipedia.org/wiki/Sphingomonas ''Sphingomonas capsulata''] in an <scene name='71/719201/Unbound_pep/1'> unbound state </scene>. Both PEPs have two domains: a catalytic binding site that forms interactions with the substrate and the propeller domain that acts as a sort of stabilizing clamp structure. <ref name="shan" /> | ||
<br>[[Image:Proteopedia article image 3.jpg|right]]<br> | <br>[[Image:Proteopedia article image 3.jpg|right]]<br> | ||
Revision as of 16:58, 7 April 2020
| |||||||||||
3D structures of gliadin
Updated on 07-April-2020
4gg6 – wGLI alpha/beta peptide + antibody + T-cell receptor – wheat
4z7u, 4z7v, 4z7w – wGLI alpha-1 peptide + antibody + T-cell receptor
4ozf, 4ozg, 4ozh, 4ozi – wGLI alpha-2 peptide + antibody + T-cell receptor
1s9v, 2nna – GLI alpha-1 peptide + antibody - synthetic
4d8p – wGLI gamma peptide + antibody
References
- ↑ Celiac Disease: MedlinePlus. Retrieved October 27, 2015, from https://www.nlm.nih.gov/medlineplus/celiacdisease.html
- ↑ Celiac Disease. Retrieved October 27, 2015, from http://www.niddk.nih.gov/health-information/health-topics/digestive-diseases/celiac-disease/Pages/facts.aspx
- ↑ Go to Science. Retrieved October 27, 2015, from http://www.sciencemag.org/content/297/5590/2275.full
- ↑ 4.0 4.1 Maiuri, L., Ciacci, C., Ricciardelli, I., Vacca, L., Raia, V., Auricchio, S., . . . Londei, M. (2003). Association between innate response to gliadin and activation of pathogenic T cells in coeliac disease. Lancet, 362(9377), 30-37. doi:10.1016/S0140-6736(03)13803-2
- ↑ Mellins, E., & Stern, L. (n.d.). HLA-DM and HLA-DO, key regulators of MHC-ll processing and presentation. Current Opinion in Immunology, 26, 115-122. February 2014. http://www.sciencedirect.com/science/article/pii/S095279151300215X
- ↑ Kim, C., Quartsen, H., Bergsen, E., Khosla, C., & Sollid, L.. (n.d.). Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease. Cross Mark, 101(12), 4175-4179. March 2004 http://www.pnas.org/content/101/12/4175.figures-only
- ↑ Henderson, K. N., Tye-Din, J. A., Reid, H. H., Chen, Z., Borg, N. A., Beissbarth, T., . . . Anderson, R. P. (2007). A structural and immunological basis for the role of human leukocyte antigen DQ8 in celiac disease. Immunity,27(1), 23-34. doi:10.1016/j.immuni.2007.05.015
- ↑ 8.0 8.1 8.2 8.3 8.4 Shan, L., I. I. Mathews, and C. Khosla. "Structural and Mechanistic Analysis of Two Prolyl nEndopeptidases: Role of Interdomain Dynamics in Catalysis and Specificity." Proceedings of the National Academy of Sciences 102.10 (2005): 3599-604. Web.
- ↑ "Permission is not required to use original figures or tables for noncommercial and educational use",Rights & Permissions, Proceedings of the National Aademy of Sciences, USA.
- ↑ Matysiak-Budnik, T., Candalh, C., Cellier, C., Dugave, C., Namane, A., Vidal-Martinez, T., . . . Heyman, M. (2005). Limited efficiency of prolyl-endopeptidase in the detoxification of gliadin peptides in celiac disease. Gastroenterology,129(3), 786-796. doi:10.1053/j.gastro.2005.06.016
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Premal Patel, Eric Martz, Alexander Berchansky, Devin Joseph, Ben Horansky, Gunnar Reiske, Katlin Cannon, Joel L. Sussman, Jaime Prilusky