We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox k11v
From Proteopedia
(Difference between revisions)
| Line 15: | Line 15: | ||
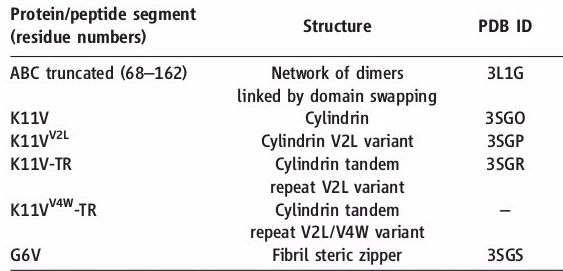
Oligomer forming segment of ABC were identified by inspection of its 3D structure and by applying the Rosetta-Profile algorithm to its sequence.Two segments of high amyloidogenic propensity,with sequences KVKVLG and GDVIEV (where D indicates Asp; E, Glu; G, Gly; I, Ile; K, Lys; and V, Val). | Oligomer forming segment of ABC were identified by inspection of its 3D structure and by applying the Rosetta-Profile algorithm to its sequence.Two segments of high amyloidogenic propensity,with sequences KVKVLG and GDVIEV (where D indicates Asp; E, Glu; G, Gly; I, Ile; K, Lys; and V, Val). | ||
| - | The entire 11-residue segment KVKVLGDVIEV forms a <scene name='77/771966/K11v_in_black/1'>hairpin loop</scene> in the 3D structure of ABC. | + | The entire 11-residue segment KVKVLGDVIEV forms a <scene name='77/771966/K11v_in_black/1'>hairpin loop</scene> in the 3D structure of ABC.The six residue segment GDVIEV ,termed <scene name='77/771966/Gdviev_black/1'>G6V</scene>,forms fibrils and microcrystals.THe microcrystals enabled us to determine the atomic structure of G6V, which proved to be a standard class 2 steric zipper, essentially an amyloid-like protofilament. |
| + | [[Image:Table_abc_first_ok.jpg]] | ||
Revision as of 14:07, 4 November 2017
Toxic Amyloid Small Oligomer’s atomic view
| |||||||||||
References
- ↑ Laganowsky A, Liu C, Sawaya MR, Whitelegge JP, Park J, Zhao M, Pensalfini A, Soriaga AB, Landau M, Teng PK, Cascio D, Glabe C, Eisenberg D. Atomic view of a toxic amyloid small oligomer. Science. 2012 Mar 9;335(6073):1228-31. PMID:22403391 doi:10.1126/science.1213151