Art:Molecular Sculpture
From Proteopedia
(→Julian Voss-Andreae) |
(→Julian Voss-Andreae) |
||
| Line 190: | Line 190: | ||
[[Image:Voss-Andreae-2008 angel of the west studio-500px.jpg]] | [[Image:Voss-Andreae-2008 angel of the west studio-500px.jpg]] | ||
</td></tr><tr><td> | </td></tr><tr><td> | ||
| - | Julian Voss-Andreae standing in his studio with his completed sculpture "Angel of the West", 2008. The core of this sculpture is a scientifically accurate rendering of an antibody molecule. Stainless steel, 12' x 12' x 4' (3.70 x 3.70 x 1.20 m). Permanent location: The Scripps Research Institute (Jupiter, FL). Used with permission<ref name="jva" />. | + | [http://julianvossandreae.com Julian Voss-Andreae] standing in his studio with his completed sculpture "Angel of the West", 2008. The core of this sculpture is a scientifically accurate rendering of an antibody molecule. Stainless steel, 12' x 12' x 4' (3.70 x 3.70 x 1.20 m). Permanent location: The Scripps Research Institute (Jupiter, FL). Used with permission<ref name="jva" />. |
</td></tr></table> | </td></tr></table> | ||
| Line 197: | Line 197: | ||
[[Image:Voss-Andreae-2003 light harvesting complex-cropped.jpg]] | [[Image:Voss-Andreae-2003 light harvesting complex-cropped.jpg]] | ||
</td></tr><tr><td> | </td></tr><tr><td> | ||
| - | ''Light Harvesting Complex'' by Julian Voss-Andreae, 2003. 22” x 25” x 25” (56 x 64 x 64 cm). Photo used with permission<ref name="jva">Permission obtained in April, 2018 from Julian Voss-Andreae.</ref>. | + | ''Light Harvesting Complex'' by [http://julianvossandreae.com Julian Voss-Andreae], 2003. 22” x 25” x 25” (56 x 64 x 64 cm). Photo used with permission<ref name="jva">Permission obtained in April, 2018 from Julian Voss-Andreae.</ref>. |
</td></tr></table> | </td></tr></table> | ||
{{Clear}} | {{Clear}} | ||
Revision as of 22:39, 12 April 2018
Physical models of macromolecules were important for scientists to understand protein structures when the first protein crystal structures were determined. From 1958 into the 1960's, these included brass wireframe models[1], facilitated by the Richards optical comparator, and smaller ball and spoke models[2]. By the early 1970's, computers were still not yet commonly capable of rendering macromolecular models (see History of Macromolecular Visualization). At that time, Byron's Bender was a popular way of making scientifically accurate protein backbone models. As computers took over scientific visualization in the 1980's and beyond, physical models became works of art as well as science. However, physical models remain very important in education[3][4], because some students learn by tactile exploration better than from a computer screen, and most students learn better when tactile exploration of physical models is available[5][6][7][8].
The scope of this article is primarily limited to sculptures and physical models of macromolecules. Some of this article was adapted from an earlier website[9].
Contents |
Beevers -> Miramodus
|
Giant model of the monomer of nylon from Miramodus.Com. Photo used with permission[10]. C H N O |
There is a strong tradition of model making in the Chemistry Department of Edinburgh University. Alexander Crum Brown, Professor of Chemistry from 1869-1908, was a pioneer in this field. In the Year the Crum Brown retired, Cecil Arnold Beevers was born in Manchester. In 1929, he became involved, under the influence of Lawrence Bragg in Manchester, in research in crystallography. With Henry Lipson, he made major contribution in the Beevers-Lipson Strips, a computational aid in calculating Fourier transforms to solve structures crystallographically.
|
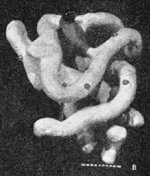
The first 3D protein structure: myoglobin at ~6 Å by Kendrew et al..
|
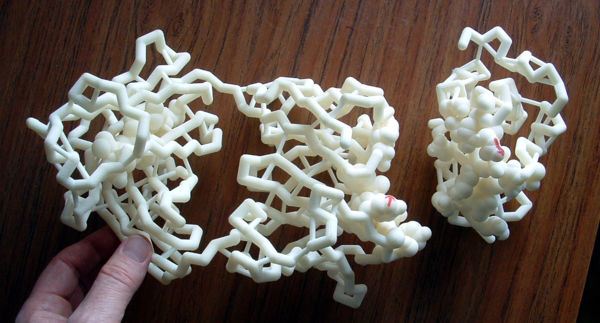
In 1958, John Kendrew (Nobel Prize 1962 with Max Perutz) and coworkers published the first structure of a protein, myoglobin[11]. It was a low resolution (6 Å) structure that looked unsatisfyingly asymmetrical. By 1961[12][13], the Kendrew group had achieved atomic resolution for myoglobin. Initially, they built brass models at a scale of 5 cm/Ångstrom. The models were built and supported within a forest of 2,500 vertical rods arranged to fill a cube six feet (2 meters) on a side. Colored clips were attached to the rods to signify electron density, and guide the building of the model. The forest of rods obscured the view of the model and made it hard to adjust. Its size made it cumbersome and problematic to move. See photo 1 and photo 2.
|
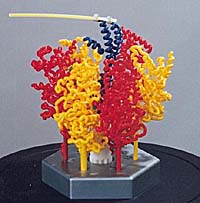
Beevers type ball and spoke model of hemoglobin at 1 cm/2 Å, available from Miramodus.Com. Hemoglobin is about 60 Å across, so the model is about 30 cm (1 ft) across. This model has one ball per residue (574 balls). It is not an all-atom model, which, if hydrogen is included, would have 8,800 atoms. Photo used with permission[10]. |
In 1965, seeking smaller and more portable models, Kendrew collaborated with Alexander A. Barker, who constructed ball and spoke models using parts provided by Beevers. These were 1 cm/Å, one fifth the scale of Kendrew's original myoglobin model, later called Beever's Miniature Models. Kendrew started to take orders for these models of myoglobin in May 1966. The price was set at £210 (roughly $600 US), a considerable sum at the time, equivalent to about $4,600 US in 2018. In all, 29 orders were received between May 1966 and March 1968 for models which were produced at the rate of one per month. SEE PHOTOS.
Beevers style ball and spoke models of proteins remain available in 2018 from Miramodus.Com. The scale for proteins is 1 cm/2 Å.
When it comes to larger sculptures, Miramodus "Giant Models" make a striking impression.
Byron Rubin
Byron's Bender
|
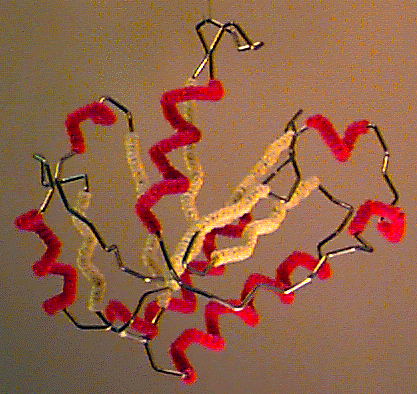
Byron's Bender wire model of the backbone trace of the I domain of CD11a (from 1lfa), an alpha/beta domain of a leukocyte integrin adhesion molecule. Alpha helices are decorated with red pipe cleaners, while beta strands are decorated yellow. Model generously constructed by Tim Herman in 1997. |
Byron Rubin, while working as a crystallographer with Jane Richardson in the early 1970's, invented a machine for bending wire to follow the backbone trace of a protein[14][15]. Kendrew-style brass wireframe models, then popular, were large and cumbersome. The small backbone wire models from Byron's Bender were the most manipulable and portable models available at the time.
An example illustrating the importance of models from Byron's Bender occurred at a scientific meeting in the mid 1970's. At this time, less than two dozen protein structures had been solved. David Davies brought a Bender model of an immunoglobulin Fab fragment, and Jane and David Richardson brought a Bender model of superoxide dismutase. While comparing these physical models at the meeting, they realized that both proteins use a similar fold, despite having only about 9% sequence identity. This incident[16] was the first recognition of the occurrence of what is now recognized as the immunoglobulin superfamily domain in proteins that are apparently unrelated by sequence.
Byron's Bender remained available through the 1990's. Tim Herman, then of the Medical College of Wisconsin (later he founded[3][4]) was one of its last avid users. Tim brought the Bender into local high schools and taught teachers and groups of students how to construct models.
Aside from the importance of the tactile as well as visual input these models provide, another of their great strengths is that they jiggle and vibrate when handled, thereby simulating thermal motion. Too often users of computer models lose sight of the fact that protein molecules in living systems are constantly flexing due to thermal motion.
(Text in this section was adapted by Eric Martz from his earlier version at History.MolviZ.Org.)
Muffler Pipe
Shortly after conceiving the idea for his Bender, crystallographer Byron Rubin realized that the machine used in Midas Muffler shops to customize automobile tailpipes operated on a similar principle, but at larger scale. He collaborated with the local shop to construct a backbone sculpture of rubredoxin about 5 feet high from stainless steel tailpipe. Rubin's rubredoxin sculpture (not shown[17]) won the Chandler competetion at the University of North Carolina in 1973, and since then has stood in the lobby of the Paul M. Gross Chemistry Building at Duke University, Durham NC USA.
MolecularSculpture.Com
|
Byron Rubin completing the installation of his sculpture of follicle stimulating hormone in 2002 at EMD-Serono, Rockland MA. Photo used with permission[18]. Photo by Eric Martz. |
| Sculpture of human neutrophil collagenase by Byron Rubin on permanent exhibition at the Smithsonian Institution, Washington DC USA. Image copyright © 2001, Byron H. Rubin, used with permission[18]. Height: 11 inches. |
In the mid-1990's, Byron Rubin resumed the creation of molecular sculptures such as the collagenase ribbon model shown at right. In his workshop near Rochester, NY, in 2001, he has constructed many beautiful protein molecular sculptures of all sizes, often for pharmaceutical companies. These can be seen at his website, MolecularSculpture.Com.
Edgar Meyer
The following text and image was submitted[9] in 2004 by Edgar Meyer (1935-2015). Citations have been added.
|
Sculpted walnut model of D-tartaric acid, the cornerstone of stereochemistry and chirality. By Edgar Meyer. |
- Early in his scientific career, Edgar Meyer made three seminal contributions to the structural sciences: the first use of color graphics for molecular modeling (1968), the initiation of the Protein Data Bank (1969-1971)[19], and the first use of networking in the life sciences (1971). These three tools[20] are the very basis of web-based structural studies so essential to the life sciences and are a source of inspiration for his artistic efforts.
- At the end of 40 years as a productive crystallographer, he spent the last 10 years [1994-2004] developing the software and hardware tools necessary to create precisely scaled models of molecules in the form of molecular sculptures. Program SCULPT reads atomic coordinates and, together with orientation and control parameters, generates “G” code for a cnc milling machine.
- Some of the sculptures are derived directly from his crystallographic studies of proteolytic and hydrolytic enzymes. The trinitatis series of sculptures comes directly from his initial observation of amazing repeats of single amino acids in proteins[21].
- These sculptures merge art and science; they invite the viewer to touch the contoured surfaces depicting atom-atom juxtapositions. The warmth and texture of noble hardwoods is the medium most frequently used for these sculptures. Commissions by the Boston Museum of Science and the Smithsonian are depicted in his web site molecular-sculpture.com.
- In addition to a variety of molecular sculptures shown there, abstractions based on atomic surface representations and juxtapositions are also illustrated. These abstractions are inspired by the wood itself.
Photographs of larger sculptures and macromolecular subjects are available[22][23][24].
Jane S. Richardson
Although crystallographer Jane Richardson did not make sculptures of protein molecules, she made an indelible impact on protein sculptors by introducing and popularizing simplified and beautiful drawings of protein backbones, notably schematic ribbon diagrams[25][26][27][28][29].
By simplifying protein models from imponderable masses of atoms to elegant ribbon diagrams, she facilitated scientists' understanding of protein folds and their families and superfamilies. Her influence can be seen in the later sculptures of Byron Rubin.
Tim Herman
|
Tim Herman in 2015 explaining secondary structure on YouTube. The Center for Biomolecular Modeling has posted many videos on YouTube. |
|
An educational, interactive model of ATPase, a molecular motor, built at the MSOE Center for BioMolecular Modeling and painted by hand. Courtesy 3D Molecular Designs. |
Tim Herman joined the faculty of the Medical College of Wisconsin in 1980. In 1998 he moved to the Milwaukee School of Engineering to establish the Center for Biomolecular Modeling, and in 1999 3DMolecularDesigns.Com. They use diverse rapid prototyping and other methods to create 3-D physical models of proteins and other molecular structures for biomedical researchers, and innovative educational models for educators. Their educational products are developed in collaboration with experienced teachers and field-tested in classrooms.
|
Antibody bound to lysozyme antigen. Large (16 x 6 in/40 x 15 cm) backbone model of the Fab fragment of a mouse IgG1k antibody bound to hen egg white lysozyme 1dkl. The epitope (on lysozyme) and paratope (on the Fab) have spacefilled atoms, enabling one to feel the shape complementarity of the binding. The catalytic cleft of the lysozyme is not occluded by Fab binding, and a tetrasaccharide substrate model is available. Some hydrogen bonds, and all disulfide bonds, are shown. Glu35 and Asp52, the key catalytic residues of lysozyme[30], are spacefilled. Nylon model courtesy Tim Herman and 3D Molecular Designs. |
Arthur Olson
Darling Models
Bathsheba Grossman
|
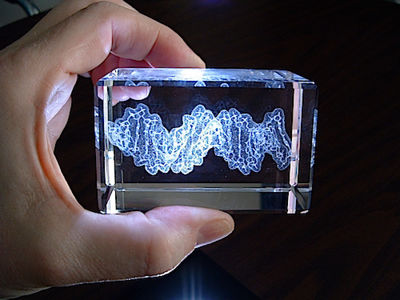
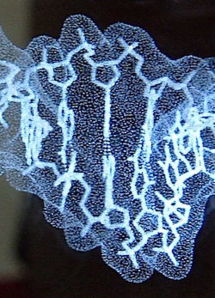
Short segment of DNA double helix (B form; based on the crystallographic result in 1au7) etched by LASER in a block of optical-quality glass by CrystalProtein.Com. | |
Sculptor Bathsheba Grossman (Bathsheba.Com) makes geometric sculptures of mathematically inspired objects from originally designed computer datasets, using rapid prototyping technologies or laser cutting of sheet metal. She also makes glass blocks containing laser-etched models of macromolecules, which she calls "Crystal Proteins" (CrystalProtein.Com). These can be designed to customer specifications. Here is a description of her methods.
See a 3D (rotatable) rendering of the DNA model at right, and click 3D Preview in her Gallery for protein and other models.
Patrick Goldsmith
Atkins, Roderick & Flammer
|
A DNA earring -- genetic jewels. Constructing these is an educational activity which provides step by step instructions. |
In 1992, as participants in Evolution and the Nature of Science Institutes (ENSI) at Indiana University, Thomas Atkins and Joyce Roderick developed an educational activity for making DNA earrings. In 2004, the project was adapted to the website by L. Flammer. A detailed lesson plan is provided along with step by step instructions for constructing DNA earrings.
Julian Voss-Andreae
|
Voss-Andreae's "Alpha Helix for Linus Pauling" (used with permission). The 10-foot (3 m) sculpture was created to honor the memory of Linus Pauling, who discovered the alpha helix in 1951. In order to emphasize the tension between the natural environment and man's image of nature's building block, the piece was powder coated in primary red, complementary in color to the green foliage embracing it. The sculpture is located in front of Linus Pauling's boyhood home 3945 SE Hawthorne Boulevard in Portland, Oregon, now home of the Linus Pauling Center for Science, Peace, and Health, which commissioned the piece. Photo used with permission[31]. |
Julian Voss-Andreae (JulianVossAndreae.Com) trained as a physicist in Europe and then as an artist in Portland, Oregon USA. His works include human forms, mathematical forms, and some inspired by protein and nucleic acid molecular structures. His construction of Angel of the West, inspired by the structures of antibody molecules, is featured in this video, along with others of his works. Voss-Andreae submitted[9] the following description in 2004:
- Voss-Andreae's current body of work plays on the sensuality and beauty which underlies sense and being itself. His work takes a literal look at the foundation of our physical existence. Voss-Andreae creates sculptures of proteins, the universal building blocks of life. More important for him than accurately copying a molecule in all its details is finding a guiding principle and following it to see whether it yields artistically interesting results. The main idea underlying all his protein sculptures is the analogy between the technique of mitered cuts and protein folding. His work resembles nature in its algorithmic quality. The former physicist calculates the cutting angles from scientific protein data using computer software he developed for that purpose. Beside the deterministic side of his work, there is an equally strong intuitive and irrational side, where his pieces stop working as scientific models and become pure art objects. Voss-Andreae's sculptures offer an emotional experience of a world that is usually accessible only through our intellect.
|
Julian Voss-Andreae standing in his studio with his completed sculpture "Angel of the West", 2008. The core of this sculpture is a scientifically accurate rendering of an antibody molecule. Stainless steel, 12' x 12' x 4' (3.70 x 3.70 x 1.20 m). Permanent location: The Scripps Research Institute (Jupiter, FL). Used with permission[31]. |
|
Light Harvesting Complex by Julian Voss-Andreae, 2003. 22” x 25” x 25” (56 x 64 x 64 cm). Photo used with permission[31]. |
Rolf Eckhardt
Mara G. Haseltine
See Also
- Byron's Bender
- History of Macromolecular Visualization
- Molecular modeling and visualization software
Notes & References
- ↑ KENDREW JC, BODO G, DINTZIS HM, PARRISH RG, WYCKOFF H, PHILLIPS DC. A three-dimensional model of the myoglobin molecule obtained by x-ray analysis. Nature. 1958 Mar 8;181(4610):662-6. PMID:13517261
- ↑ A. A. Barker's Models of Myoglobin at the History of Visualization of Biological Macromolecules (History.MolviZ.Org).
- ↑ 3.0 3.1 3dMolecularDesigns.Com
- ↑ 4.0 4.1 Center for Biomolecular Modeling, Milwaukee School of Engineering.
- ↑ Roberts JR, Hagedorn E, Dillenburg P, Patrick M, Herman T. Physical models enhance molecular three-dimensional literacy in an introductory biochemistry course*. Biochem Mol Biol Educ. 2005 Mar;33(2):105-10. doi: 10.1002/bmb.2005.494033022426. PMID:21638554 doi:http://dx.doi.org/10.1002/bmb.2005.494033022426
- ↑ Bain, G., Yi, J., Beikmohamadi, M., Herman, T. and Patrick, M. 2006. Using physical models to teach concepts of biochemical structure and structure depiction in the introductory chemistry laboratory. J. Chem. Educ. 83(9):1322-1324.
- ↑ Herman T, Morris J, Colton S, Batiza A, Patrick M, Franzen M, Goodsell DS. Tactile teaching: Exploring protein structure/function using physical models*. Biochem Mol Biol Educ. 2006 Jul;34(4):247-54. doi: 10.1002/bmb.2006.494034042649. PMID:21638686 doi:http://dx.doi.org/10.1002/bmb.2006.494034042649
- ↑ Harris MA, Peck RF, Colton S, Morris J, Chaibub Neto E, Kallio J. A combination of hand-held models and computer imaging programs helps students answer oral questions about molecular structure and function: a controlled investigation of student learning. CBE Life Sci Educ. 2009 Spring;8(1):29-43. doi: 10.1187/cbe.08-07-0039. PMID:19255134 doi:http://dx.doi.org/10.1187/cbe.08-07-0039
- ↑ 9.0 9.1 9.2 Much of the content of this article on Molecular Sculpture was adapted by Eric Martz from a section of the same title in his earlier project, the World Index of BioMolecular Visualization Resources. That project went off-line, inadvertantly, in 2012.
- ↑ 10.0 10.1 Permission obtained in April, 2018 from Gavin Whittaker at Miramodus.Com.
- ↑ KENDREW JC, BODO G, DINTZIS HM, PARRISH RG, WYCKOFF H, PHILLIPS DC. A three-dimensional model of the myoglobin molecule obtained by x-ray analysis. Nature. 1958 Mar 8;181(4610):662-6. PMID:13517261
- ↑ KENDREW JC, DICKERSON RE, STRANDBERG BE, HART RG, DAVIES DR, PHILLIPS DC, SHORE VC. Structure of myoglobin: A three-dimensional Fourier synthesis at 2 A. resolution. Nature. 1960 Feb 13;185(4711):422-7. PMID:18990802
- ↑ KENDREW JC, WATSON HC, STRANDBERG BE, DICKERSON RE, PHILLIPS DC, SHORE VC. The amino-acid sequence x-ray methods, and its correlation with chemical data. Nature. 1961 May 20;190:666-70. PMID:13752474
- ↑ Rubin, Byron; Richardson Jane S. The simple construction of protein alpha-carbon models. Biopolymers. 1972; 11(11):2381-5. PDF
- ↑ Rubin, Byron. 1985. Macromolecule backbone models. Methods in Enzymology 115:391-7.
- ↑ Richardson JS, Richardson DC, Thomas KA, Silverton EW, Davies DR. Similarity of three-dimensional structure between the immunoglobulin domain and the copper, zinc superoxide dismutase subunit. J Mol Biol. 1976 Apr 5;102(2):221-35. PMID:1271464 doi:http://dx.doi.org/10.1016/S0022-2836(76)80050-2
- ↑ We have been unable to obtain a photograph of this sculpture. If you can provide one, please send it to
 .
. - ↑ 18.0 18.1 Permission obtained in April, 2018 from Byron Rubin
- ↑ Meyer EF. The first years of the Protein Data Bank. Protein Sci. 1997 Jul;6(7):1591-7. doi: 10.1002/pro.5560060724. PMID:9232661 doi:http://dx.doi.org/10.1002/pro.5560060724
- ↑ Memoir by Edgar F. Meyer, American Crystallographic Association History
- ↑ WWWWhy does nature stutter? A survey of strands of repeated amino acids. Edgar F. Meyer & W. John Tollett, Jr. Acta Cryst. D57, 181-186, 2004. doi.org/10.1107/S0907444900018023.
- ↑ Obituary, Edgar Meyer (1935 - 2015), American Crystallographic Association.
- ↑ Edgar Meyer's favorite structure is: co-chaperonin, Chemical & Engineering News
- ↑ Monumental Molecules by Edgar Meyer, International Union of Crystallography Newsletter 20(3), 2012.
- ↑ Richardson JS, Tainer JA, Richardson DC. An illustrated museum of protein structures. Biophys J. 1980 Oct;32(1):211-3. doi: 10.1016/S0006-3495(80)84934-4. PMID:19431356 doi:http://dx.doi.org/10.1016/S0006-3495(80)84934-4
- ↑ Richardson JS. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167-339. PMID:7020376
- ↑ Richardson JS. Describing patterns of protein tertiary structure. Methods Enzymol. 1985;115:341-58. PMID:4079792
- ↑ Richardson JS. Schematic drawings of protein structures. Methods Enzymol. 1985;115:359-80. PMID:3853075
- ↑ Richardson JS, Richardson DC, Tweedy NB, Gernert KM, Quinn TP, Hecht MH, Erickson BW, Yan Y, McClain RD, Donlan ME, et al.. Looking at proteins: representations, folding, packing, and design. Biophysical Society National Lecture, 1992. Biophys J. 1992 Nov;63(5):1185-209. PMID:1477272
- ↑ Vocadlo DJ, Davies GJ, Laine R, Withers SG. Catalysis by hen egg-white lysozyme proceeds via a covalent intermediate. Nature. 2001 Aug 23;412(6849):835-8. PMID:11518970 doi:10.1038/35090602
- ↑ 31.0 31.1 31.2 Permission obtained in April, 2018 from Julian Voss-Andreae.