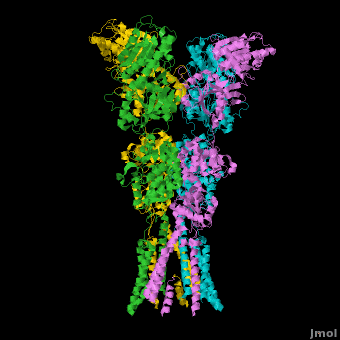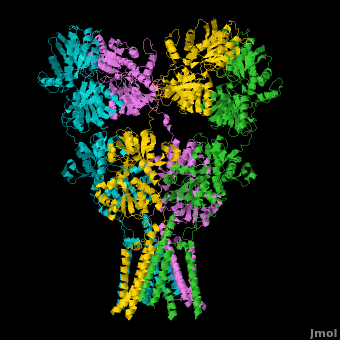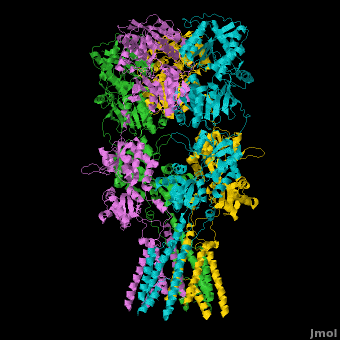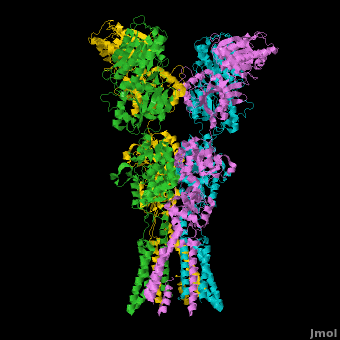Ionotropic Glutamate Receptors
From Proteopedia
(Difference between revisions)
| Line 69: | Line 69: | ||
*Ionotropic glutamate receptor 2 | *Ionotropic glutamate receptor 2 | ||
| - | **[[4u1y]], [[4u2p]], [[5l1b]] - rIGluR2 <br /> | ||
| - | **[[3n6v]], [[3o2j]], [[5n6p]] - rIGluR2 ATD (mutant) – rat<br /> | ||
| - | **[[3hsy]], [[3h5v]], [[3h5w]] - rIGluR2 ATD<br /> | ||
**[[2wjw]], [[2wjx]] – hIGluR2 ATD - human<br /> | **[[2wjw]], [[2wjx]] – hIGluR2 ATD - human<br /> | ||
**[[3rn8]], [[3rnn]] – hIGluR2 LBD + potentiator<br /> | **[[3rn8]], [[3rnn]] – hIGluR2 LBD + potentiator<br /> | ||
**[[5ybg]], [[5ybf]] - hIGluR2 LBD + potentiator + Glu<br /> | **[[5ybg]], [[5ybf]] - hIGluR2 LBD + potentiator + Glu<br /> | ||
| + | **[[4u1y]], [[4u2p]], [[5l1b]] - rIGluR2 <br /> | ||
| + | **[[3n6v]], [[3o2j]], [[5n6p]] - rIGluR2 ATD (mutant) – rat<br /> | ||
| + | **[[3hsy]], [[3h5v]], [[3h5w]] - rIGluR2 ATD<br /> | ||
**[[3bki]], [[5l1e]], [[5l1f]] [[5l1e]], [[5l1f]], [[5l1g]], [[5l1h]] - rIGluR2 LBD + inhibitor<br /> | **[[3bki]], [[5l1e]], [[5l1f]] [[5l1e]], [[5l1f]], [[5l1g]], [[5l1h]] - rIGluR2 LBD + inhibitor<br /> | ||
**[[1fto]], [[4h8j]] - rIGluR2 LBD<br /> | **[[1fto]], [[4h8j]] - rIGluR2 LBD<br /> | ||
| Line 127: | Line 127: | ||
*Ionotropic glutamate receptor δ | *Ionotropic glutamate receptor δ | ||
| + | **[[5kc8]] – hIGluRδ2 ATD<br /> | ||
**[[5kc9]] – IGluRδ1 C-terminal - mouse <br /> | **[[5kc9]] – IGluRδ1 C-terminal - mouse <br /> | ||
**[[5l2e]] – rIGluRδ2 <br /> | **[[5l2e]] – rIGluRδ2 <br /> | ||
| - | **[[5kc8]] – hIGluRδ2 ATD<br /> | ||
**[[5cc2]] – rIGluRδ2 LBD + agonist<br /> | **[[5cc2]] – rIGluRδ2 LBD + agonist<br /> | ||
| Line 166: | Line 166: | ||
**[[3fuz]], [[2zns]] – hIGluR5 LBD + Glu<br /> | **[[3fuz]], [[2zns]] – hIGluR5 LBD + Glu<br /> | ||
| + | **[[3fv1]], [[3fv2]], [[3fvg]], [[3fvk]], [[3fvn]], [[3fvo]], [[2znt]], [[2znu]] - hIGluR5 LBD + agonist<br /> | ||
**[[1txf]] - rIGluR5 LBD + Glu<br /> | **[[1txf]] - rIGluR5 LBD + Glu<br /> | ||
**[[2ojt]] - rIGluR5 + anion<br /> | **[[2ojt]] - rIGluR5 + anion<br /> | ||
| - | **[[3fv1]], [[3fv2]], [[3fvg]], [[3fvk]], [[3fvn]], [[3fvo]], [[2znt]], [[2znu]] - hIGluR5 LBD + agonist<br /> | ||
**[[3c31]], [[3c32]], [[3c33]], [[3c34]], [[3c35]], [[3c36]] - rIGluR5 LBD + ion<br /> | **[[3c31]], [[3c32]], [[3c33]], [[3c34]], [[3c35]], [[3c36]] - rIGluR5 LBD + ion<br /> | ||
**[[2wky]], [[3gba]], [[3gbb]], [[2pbw]], [[2f34]], [[2f35]], [[2f36]] – rIGluR5 LBD + agonist<br /> | **[[2wky]], [[3gba]], [[3gbb]], [[2pbw]], [[2f34]], [[2f35]], [[2f36]] – rIGluR5 LBD + agonist<br /> | ||
| Line 194: | Line 194: | ||
*Ionotropic kainate receptor 2 | *Ionotropic kainate receptor 2 | ||
| + | **[[3qxm]] - hGluK2 LBD + toxin<br /> | ||
**[[2xxr]] – rGluK2 LBD + Glu<br /> | **[[2xxr]] – rGluK2 LBD + Glu<br /> | ||
**[[4h8i]], [[5cmm]] - rGluK2 LBD + Glu derivative<br /> | **[[4h8i]], [[5cmm]] - rGluK2 LBD + Glu derivative<br /> | ||
| Line 204: | Line 205: | ||
**[[5kuh]] - rGluK2 LBD (mutant) + agonist<br /> | **[[5kuh]] - rGluK2 LBD (mutant) + agonist<br /> | ||
**[[4uqq]] - rGluK2 ATD,LBD,TMD (mutant) + agonist<br /> | **[[4uqq]] - rGluK2 ATD,LBD,TMD (mutant) + agonist<br /> | ||
| - | **[[3qxm]] - hGluK2 LBD + toxin | ||
*Ionotropic kainate receptor 3 | *Ionotropic kainate receptor 3 | ||
| Line 230: | Line 230: | ||
**[[4kcc]] – rNMDAR LBD <br /> | **[[4kcc]] – rNMDAR LBD <br /> | ||
**[[3q41]] – rNMDAR N ATD <br /> | **[[3q41]] – rNMDAR N ATD <br /> | ||
| - | **[[3qek]] – XlNMDA R ATD (mutant) – ''Xenopus laevis''<br /> | ||
**[[1y1m]], [[1y1z]], [[1y20]] – NMDAR LBD (mutant) + partial agonist<br /> | **[[1y1m]], [[1y1z]], [[1y20]] – NMDAR LBD (mutant) + partial agonist<br /> | ||
**[[2hqw]] – NMDAR C terminal + calmodulin<br /> | **[[2hqw]] – NMDAR C terminal + calmodulin<br /> | ||
| Line 238: | Line 237: | ||
**[[4kfq]] – rNMDAR LBD + antagonist<br /> | **[[4kfq]] – rNMDAR LBD + antagonist<br /> | ||
**[[5fxk]], [[5fxi]], [[5fxh]], [[5fxg]] – rNMDAR (mutant) – Cryo EM<br /> | **[[5fxk]], [[5fxi]], [[5fxh]], [[5fxg]] – rNMDAR (mutant) – Cryo EM<br /> | ||
| + | **[[3qek]] – XlNMDA R ATD (mutant) – ''Xenopus laevis''<br /> | ||
*''NMDAR subunit NR2A'' | *''NMDAR subunit NR2A'' | ||
| Line 269: | Line 269: | ||
*''NMDAR subunit NR1+NR2A'' | *''NMDAR subunit NR1+NR2A'' | ||
| + | **[[5h8f]] – hNMDAR LBD <br /> | ||
| + | **[[5tpa]], [[5tp9]], [[5kdt]], [[5kcj]], [[5i2n]], [[5i2k]], [[5h8q]], [[5h8n]], [[5h8h]] – hNMDAR LBD + allosteric modulator<br /> | ||
**[[4nf4]], [[4nf5]], [[4nf6]], [[4nf8]], [[2a5t]] – rNMDAR LBD <br /> | **[[4nf4]], [[4nf5]], [[4nf6]], [[4nf8]], [[2a5t]] – rNMDAR LBD <br /> | ||
| - | **[[5h8f]] – hNMDAR LBD <br /> | ||
**[[5vij]], [[5vii]], [[5vih]], [[5dex]], [[5u8c]] – rNMDAR LBD + antagonist<br /> | **[[5vij]], [[5vii]], [[5vih]], [[5dex]], [[5u8c]] – rNMDAR LBD + antagonist<br /> | ||
**[[5jty]], [[5fxj]] – rNMDAR + agonist<br /> | **[[5jty]], [[5fxj]] – rNMDAR + agonist<br /> | ||
**[[5tq0]], [[5tq2]] – rNMDAR LBD + antibody<br /> | **[[5tq0]], [[5tq2]] – rNMDAR LBD + antibody<br /> | ||
| + | **[[5i59]], [[5i58]], [[5i57]], [[5i56]] – rNMDAR LBD + allosteric modulator<br /> | ||
**[[5tpw]] – XlNMDAR1 LBD (mutant) + rNMDAR2A LBD + antibody<br /> | **[[5tpw]] – XlNMDAR1 LBD (mutant) + rNMDAR2A LBD + antibody<br /> | ||
**[[5ewm]], [[5ewl]] – XlNMDAR1 ATD (mutant) + rNMDAR2A ATD (mutant) + antagonist<br /> | **[[5ewm]], [[5ewl]] – XlNMDAR1 ATD (mutant) + rNMDAR2A ATD (mutant) + antagonist<br /> | ||
| - | **[[5tpa]], [[5tp9]], [[5kdt]], [[5kcj]], [[5i2n]], [[5i2k]], [[5h8q]], [[5h8n]], [[5h8h]] – hNMDAR LBD + allosteric modulator<br /> | ||
| - | **[[5i59]], [[5i58]], [[5i57]], [[5i56]] – rNMDAR LBD + allosteric modulator<br /> | ||
*''NMDAR subunit NR1+NR2B'' | *''NMDAR subunit NR1+NR2B'' | ||
| Line 283: | Line 283: | ||
**[[5tpz]] – rNMDAR LBD <br /> | **[[5tpz]] – rNMDAR LBD <br /> | ||
**[[4pe5]], [[4tll]], [[4tlm]] – rNMDAR (mutants)<br /> | **[[4pe5]], [[4tll]], [[4tlm]] – rNMDAR (mutants)<br /> | ||
| - | **[[3qel]], [[3qem]], [[5ewj]] – XlNMDAR ATD (mutant) + ifenprodil<br /> | ||
**[[5b3j]] – XlNMDAR1 ATD (mutant) + rNMDAR2B ATD (mutant) + antibody<br /> | **[[5b3j]] – XlNMDAR1 ATD (mutant) + rNMDAR2B ATD (mutant) + antibody<br /> | ||
**[[5ipv]], [[5ipt]], [[5ips]], [[5ipr]], [[5ipq]], [[5iov]], [[5iou]] – rNMDAR2B (mutant) + NMDAR1 + inhibitor<br /> | **[[5ipv]], [[5ipt]], [[5ips]], [[5ipr]], [[5ipq]], [[5iov]], [[5iou]] – rNMDAR2B (mutant) + NMDAR1 + inhibitor<br /> | ||
**[[5ipu]] – XlNMDAR2B (mutant) + NMDAR1 (mutant) + inhibitor<br /> | **[[5ipu]] – XlNMDAR2B (mutant) + NMDAR1 (mutant) + inhibitor<br /> | ||
| - | + | **[[3qel]], [[3qem]], [[5ewj]] – XlNMDAR ATD (mutant) + ifenprodil<br /> | |
}} | }} | ||
==Topic Page on Glutamate Receptor GluA2 structure== | ==Topic Page on Glutamate Receptor GluA2 structure== | ||
Revision as of 09:18, 11 July 2019
| |||||||||||
Page Development
This article was developed based on lectures given in Chemistry 543 by Prof. Clarence E. Schutt at Princeton University.
3D structures of glutamate receptor
Updated on 11-July-2019
A topic page describing in detail the GluA2 structure described in 3kg2
Topic Page on Glutamate Receptor GluA2 structure
There is a topic page describing in detail the GluA2 structure described in 3kg2. The page is meant to complement the original publication of the structure by Sobolevsky et al.[2][9] with matching colors, etc..
See Also
- Glutamate Receptor Symmetry Analysis
- Glutamate receptor (GluA2) structure in detail
- Metabotropic glutamate receptor
- Membrane Channels & Pumps
- Alzheimer's Disease
References
- ↑ 1.0 1.1 1.2 Jin R, Clark S, Weeks AM, Dudman JT, Gouaux E, Partin KM. Mechanism of positive allosteric modulators acting on AMPA receptors. J Neurosci. 2005 Sep 28;25(39):9027-36. PMID:16192394 doi:25/39/9027
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009 Dec 10;462(7274):745-56. Epub . PMID:19946266 doi:10.1038/nature08624
- ↑ 3.0 3.1 3.2 3.3 Purcell AE, Jeon OH, Zimmerman AW, Blue ME, Pevsner J. Postmortem brain abnormalities of the glutamate neurotransmitter system in autism. Neurology. 2001 Nov 13;57(9):1618-28. PMID:11706102
- ↑ Welsh JP, Ahn ES, Placantonakis DG. Is autism due to brain desynchronization? Int J Dev Neurosci. 2005 Apr-May;23(2-3):253-63. PMID:15749250 doi:10.1016/j.ijdevneu.2004.09.002
- ↑ Zuo J, De Jager PL, Takahashi KA, Jiang W, Linden DJ, Heintz N. Neurodegeneration in Lurcher mice caused by mutation in delta2 glutamate receptor gene. Nature. 1997 Aug 21;388(6644):769-73. PMID:9285588 doi:10.1038/42009
- ↑ Rubenstein JL, Merzenich MM. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2003 Oct;2(5):255-67. PMID:14606691
- ↑ Jin R, Singh SK, Gu S, Furukawa H, Sobolevsky AI, Zhou J, Jin Y, Gouaux E. Crystal structure and association behaviour of the GluR2 amino-terminal domain. EMBO J. 2009 Jun 17;28(12):1812-23. Epub 2009 May 21. PMID:19461580 doi:10.1038/emboj.2009.140
- ↑ HERRMANN GR, HEJTMANCIK MR, GRAHAM RN, MARBURGER RC. A new superior oral diuretic drug, chlorothiazide (diuril); clinical evaluation. Tex State J Med. 1958 Sep;54(9):639-45. PMID:13580922
- ↑ Wollmuth LP, Traynelis SF. Neuroscience: Excitatory view of a receptor. Nature. 2009 Dec 10;462(7274):729-31. PMID:20010675 doi:10.1038/462729a
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, David Canner, Wayne Decatur, Alexander Berchansky, Joel L. Sussman