This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
CRISPR-Cas9
From Proteopedia
(Difference between revisions)
| Line 74: | Line 74: | ||
'''Structural Basis for the Orthogonal Recognition of sgRNA Scaffolds''' | '''Structural Basis for the Orthogonal Recognition of sgRNA Scaffolds''' | ||
| - | A comparison of the quaternary complex structures of SaCas9 | + | A comparison of the quaternary complex structures of SaCas9 and SpCas9 revealed that the structurally diverse REC and WED domains recognize distinct structural features of the repeat:anti-repeat duplex, allowing the cognate sgRNAs to be distinguished in a highly specific manner. The SaCas9 WED domain has a new fold comprising a twisted five-stranded β-sheet flanked by four α-helices, and is responsible for the recognition of the distorted repeat:anti-repeat duplex, as described above. In contrast, the SpCas9 WED domain adopts a compact loop conformation and interacts with the repeat:anti-repeat duplex, which is structurally different from that of the SaCas9 sgRNA. The AnCas9 WED domain has yet another different fold containing three antiparallel b-hairpins. These structural differences in the WED domains are consistent with the variations in the sgRNA scaffolds among the CRISPR-Cas9 systems. The REC lobes also contribute to the orthogonal recognition of the sgRNA scaffolds. While the REC lobes of SaCas9 and SpCas9 share structural similarity, the SpCas9 REC lobe has four characteristic insertions (Ins 1–4), which are absent in the SaCas9 REC lobe. Ins 2 (also known as the REC2 domain) does not contact the nucleic acids in the SpCas9 structures and is dispensable for the DNA cleavage activity, consistent with the absence of Ins 2 in SaCas9 (Figures 4E and 4F). Ins 1 and 3 recognize the SpCas9-specific internal loop in the repeat:anti-repeat duplex (Figure 4F and Figure S6C). In particular, Ins 3 interacts with the flipped-out G43 and U44 in the repeat:anti-repeat duplex in base-specific manners (Figure S6C). In addition, Ins 4 interacts with stem loop 1 of the SpCas9 sgRNA, which is shorter than that of the SaCas9 sgRNA. Together, these structural observations demonstrate how the Cas9 orthologs recognize their cognate sgRNAs in orthogonal manners, using specific combinations of the structurally diverse REC and WED domains. |
| - | and SpCas9 revealed that the structurally diverse REC and WED domains recognize distinct structural | + | |
| - | features of the repeat:anti-repeat | + | |
| - | duplex, allowing the cognate sgRNAs to | + | |
| - | be distinguished in a highly specific | + | |
| - | manner | + | |
| - | + | ||
| - | new fold comprising a twisted | + | |
| - | + | ||
| - | helices, and is responsible for the recognition | + | |
| - | of the distorted repeat:anti-repeat | + | |
| - | duplex, as described above | + | |
| - | + | ||
| - | contrast, the SpCas9 WED domain | + | |
| - | adopts a compact loop conformation | + | |
| - | and interacts with the repeat:anti-repeat | + | |
| - | duplex, which is structurally different | + | |
| - | from that of the SaCas9 sgRNA | + | |
| - | + | ||
| - | + | ||
| - | The AnCas9 WED domain has yet | + | |
| - | another different fold containing three | + | |
| - | antiparallel b-hairpins | + | |
| - | + | ||
| - | in the WED domains are consistent with | + | |
| - | the variations in the sgRNA scaffolds | + | |
| - | among the CRISPR-Cas9 systems | + | |
| - | + | ||
| - | + | ||
| - | orthogonal recognition of the sgRNA | + | |
| - | scaffolds. While the REC lobes of | + | |
| - | SaCas9 and SpCas9 share structural | + | |
| - | similarity, the SpCas9 REC lobe has | + | |
| - | four characteristic insertions (Ins 1–4), | + | |
| - | which are absent in the SaCas9 REC | + | |
| - | lobe | + | |
| - | known as the REC2 domain) does not contact the nucleic acids | + | |
| - | in the SpCas9 structures and is dispensable for the DNA cleavage | + | |
| - | activity | + | |
| - | of Ins 2 in SaCas9 (Figures 4E and 4F). Ins 1 and 3 recognize the | + | |
| - | SpCas9-specific internal loop in the repeat:anti-repeat duplex | + | |
| - | (Figure 4F and Figure S6C). In particular, Ins 3 interacts with | + | |
| - | the flipped-out G43 and U44 in the repeat:anti-repeat duplex in | + | |
| - | base-specific manners (Figure S6C). In addition, Ins 4 interacts | + | |
| - | with stem loop 1 of the SpCas9 sgRNA, which is shorter than | + | |
| - | that of the SaCas9 sgRNA | + | |
| - | + | ||
| - | how the Cas9 orthologs recognize their cognate sgRNAs in | + | |
| - | orthogonal manners, using specific combinations of the structurally | + | |
| - | diverse REC and WED domains. | + | |
=See aslo= | =See aslo= | ||
*[[Cas9]] | *[[Cas9]] | ||
Revision as of 16:14, 26 August 2018
| |||||||||||
References
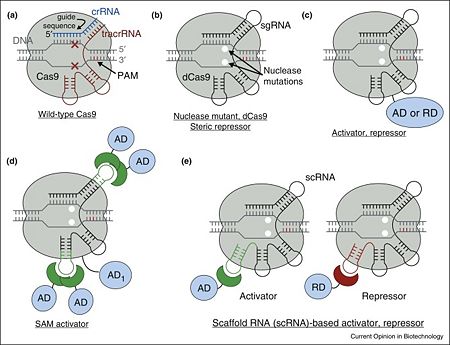
- ↑ 1.0 1.1 1.2 1.3 1.4 Didovyk A, Borek B, Tsimring L, Hasty J. Transcriptional regulation with CRISPR-Cas9: principles, advances, and applications. Curr Opin Biotechnol. 2016 Aug;40:177-84. doi: 10.1016/j.copbio.2016.06.003. Epub, 2016 Jun 23. PMID:27344519 doi:http://dx.doi.org/10.1016/j.copbio.2016.06.003
- ↑ Brophy JA, Voigt CA. Principles of genetic circuit design. Nat Methods. 2014 May;11(5):508-20. doi: 10.1038/nmeth.2926. PMID:24781324 doi:http://dx.doi.org/10.1038/nmeth.2926
- ↑ Straubeta A, Lahaye T. Zinc fingers, TAL effectors, or Cas9-based DNA binding proteins: what's best for targeting desired genome loci? Mol Plant. 2013 Sep;6(5):1384-7. doi: 10.1093/mp/sst075. Epub 2013 May 29. PMID:23718948 doi:http://dx.doi.org/10.1093/mp/sst075
- ↑ Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014 Apr;32(4):347-55. doi: 10.1038/nbt.2842. Epub 2014 Mar 2. PMID:24584096 doi:http://dx.doi.org/10.1038/nbt.2842
- ↑ Marraffini LA. CRISPR-Cas immunity in prokaryotes. Nature. 2015 Oct 1;526(7571):55-61. doi: 10.1038/nature15386. PMID:26432244 doi:http://dx.doi.org/10.1038/nature15386
- ↑ Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJ, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015 Nov;13(11):722-36. doi: 10.1038/nrmicro3569. Epub 2015, Sep 28. PMID:26411297 doi:http://dx.doi.org/10.1038/nrmicro3569
- ↑ 7.0 7.1 7.2 7.3 Jiang F, Zhou K, Ma L, Gressel S, Doudna JA. STRUCTURAL BIOLOGY. A Cas9-guide RNA complex preorganized for target DNA recognition. Science. 2015 Jun 26;348(6242):1477-81. doi: 10.1126/science.aab1452. PMID:26113724 doi:http://dx.doi.org/10.1126/science.aab1452
- ↑ 8.0 8.1 8.2 8.3 8.4 Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012 Aug 17;337(6096):816-21. doi: 10.1126/science.1225829. Epub 2012, Jun 28. PMID:22745249 doi:http://dx.doi.org/10.1126/science.1225829
- ↑ 9.0 9.1 9.2 Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012 Sep 25;109(39):E2579-86. Epub 2012 Sep 4. PMID:22949671 doi:http://dx.doi.org/10.1073/pnas.1208507109
- ↑ 10.0 10.1 10.2 10.3 Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013 Feb 28;152(5):1173-83. doi: 10.1016/j.cell.2013.02.022. PMID:23452860 doi:http://dx.doi.org/10.1016/j.cell.2013.02.022
- ↑ 11.0 11.1 11.2 Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013 Aug;41(15):7429-37. doi: 10.1093/nar/gkt520. Epub 2013, Jun 12. PMID:23761437 doi:http://dx.doi.org/10.1093/nar/gkt520
- ↑ Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014 Jul;32(7):677-83. doi: 10.1038/nbt.2916. Epub 2014 May 18. PMID:24837660 doi:http://dx.doi.org/10.1038/nbt.2916
- ↑ Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, Kaplan M, Iavarone AT, Charpentier E, Nogales E, Doudna JA. Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation. Science. 2014 Feb 6. PMID:24505130 doi:http://dx.doi.org/10.1126/science.1247997
- ↑ Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014 Feb 27;156(5):935-49. doi: 10.1016/j.cell.2014.02.001. Epub 2014 Feb, 13. PMID:24529477 doi:http://dx.doi.org/10.1016/j.cell.2014.02.001
- ↑ Jiang F, Taylor DW, Chen JS, Kornfeld JE, Zhou K, Thompson AJ, Nogales E, Doudna JA. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science. 2016 Jan 14. pii: aad8282. PMID:26841432 doi:http://dx.doi.org/10.1126/science.aad8282
- ↑ Wei Y, Terns RM, Terns MP. Cas9 function and host genome sampling in Type II-A CRISPR-Cas adaptation. Genes Dev. 2015 Feb 15;29(4):356-61. doi: 10.1101/gad.257550.114. PMID:25691466 doi:http://dx.doi.org/10.1101/gad.257550.114
- ↑ 17.0 17.1 17.2 Heler R, Samai P, Modell JW, Weiner C, Goldberg GW, Bikard D, Marraffini LA. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature. 2015 Mar 12;519(7542):199-202. doi: 10.1038/nature14245. Epub 2015 Feb, 18. PMID:25707807 doi:http://dx.doi.org/10.1038/nature14245
- ↑ 18.0 18.1 Nielsen AA, Voigt CA. Multi-input CRISPR/Cas genetic circuits that interface host regulatory networks. Mol Syst Biol. 2014 Nov 24;10:763. doi: 10.15252/msb.20145735. PMID:25422271
- ↑ 19.0 19.1 Didovyk A, Borek B, Hasty J, Tsimring L. Orthogonal Modular Gene Repression in Escherichia coli Using Engineered CRISPR/Cas9. ACS Synth Biol. 2016 Jan 15;5(1):81-8. doi: 10.1021/acssynbio.5b00147. Epub 2015 , Sep 30. PMID:26390083 doi:http://dx.doi.org/10.1021/acssynbio.5b00147
- ↑ 20.0 20.1 Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013 Jul 18;154(2):442-51. doi: 10.1016/j.cell.2013.06.044. Epub 2013 Jul, 11. PMID:23849981 doi:http://dx.doi.org/10.1016/j.cell.2013.06.044
- ↑ 21.0 21.1 Farzadfard F, Perli SD, Lu TK. Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS Synth Biol. 2013 Oct 18;2(10):604-13. doi: 10.1021/sb400081r. Epub 2013 Sep, 11. PMID:23977949 doi:http://dx.doi.org/10.1021/sb400081r
- ↑ 22.0 22.1 Kiani S, Beal J, Ebrahimkhani MR, Huh J, Hall RN, Xie Z, Li Y, Weiss R. CRISPR transcriptional repression devices and layered circuits in mammalian cells. Nat Methods. 2014 Jul;11(7):723-6. doi: 10.1038/nmeth.2969. Epub 2014 May 5. PMID:24797424 doi:http://dx.doi.org/10.1038/nmeth.2969
- ↑ Nishimasu H, Cong L, Yan WX, Ran FA, Zetsche B, Li Y, Kurabayashi A, Ishitani R, Zhang F, Nureki O. Crystal Structure of Staphylococcus aureus Cas9. Cell. 2015 Aug 27;162(5):1113-26. doi: 10.1016/j.cell.2015.08.007. PMID:26317473 doi:http://dx.doi.org/10.1016/j.cell.2015.08.007