SEE ALSO CRISPR-Cas
Background
Highlights
- CRISPR-Cas9 is a powerful tool to modulate transcription in wide range of cell types.
- An expanding set of CRISPR-based transcription effectors is available.
- Gene networks can be efficiently probed and modified for biotechnology applications.[1]
CRISPR-Cas9 has recently emerged as a promising system for multiplexed genome editing as well as epigenome and transcriptome perturbation. Due to its specificity, ease of use and highly modular programmable nature, it has been widely adopted for a variety of applications such as genome editing, transcriptional inhibition and activation, genetic screening, DNA localization imaging, and many more. In this review, we will discuss non-editing applications of CRISPR-Cas9 for transcriptome perturbation, metabolic engineering, and synthetic biology.[1]
Since the early days of genetic engineering there has been a need for control of gene expression. Naturally occurring transcription factors (TFs) have traditionally been used to achieve this goal (reviewed in [2]). However, their limited DNA binding sequence space required installing specific sequences within the transcription regulatory elements of the target genes. This can be technically difficult and may have unintended consequences on gene expression. Zinc fingers (ZFs) and transcription activator-like effectors (TALEs) were developed to overcome the fixed binding sequence requirements of native TFs. However, both ZFs and TALEs have significant limitations. ZFs have complicated design criteria and large highly repetitive TALE genes are difficult to synthesize and clone (reviewed in [3][4]). These challenges have recently been overcome using CRISPR-Cas9 based TFs. The biochemical properties of CRISPR-Cas9 based TFs that enable such flexibility and describe their applications to synthetic gene circuit design and multi-plexed perturbation of native gene networks.[1]
Transcriptional regulation with CRISPR-Cas9
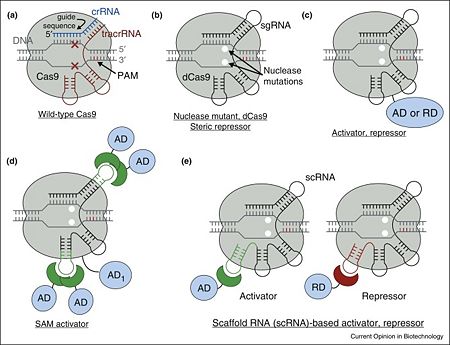
Figure 1. Overview of Cas9 nuclease and dCas9-based transcription factors. (a) Wild-type Cas9 endonuclease guided by crRNA:tracrRNA to a specific site in DNA creates a double-stranded DNA break. (b) dCas9, nuclease deactivated mutant of Cas9, is an RNA programmable DNA binding protein. It can act as a steric repressor of transcription in prokaryotes and eukaryotes. sgRNA is an artificial chimeric molecule consisting of crRNA and tracrRNA molecules connected with a short loop. (c) dCas9 fusion with various transcription effectors can be used to repress or activate transcription. (d) Effector domains can be recruited by sgRNA in addition to dCas9 for enhanced activity. (e) sgRNA can be modified with specific protein binding hairpins to concurrently recruit repressor or activator domains in the same cell.
[1] Cas9 is a key protein of bacterial Type II CRISPR adaptive immune system (reviewed in [5]). In its native context, Cas9 is an RNA-guided endonuclease that is responsible for targeted degradation of the invading foreign DNA–plasmids and phages. Cas9 is directed to its DNA targets by forming a ribonucleoprotein complex with two small non-coding RNAs: CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA) (Figure 1a). In the less common class 2 CRISPR-Cas systems (types II, V, and VI), which are almost completely restricted to bacteria, the effector complex is represented by a single multidomain protein [6]. The best-characterized class 2 effector is Cas9 (type II), the RNA-dependent endonuclease that contains two unrelated nuclease domains, HNH and RuvC, that are responsible for the cleavage of the target and the displaced strand, respectively, in the crRNA–target DNA complex (, 4zt0). The type II loci also encode a trans-acting CRISPR RNA (tracrRNA) that evolved from the corresponding CRISPR repeat and is essential for pre-crRNA processing and target recognition in type II systems. Cas9 is directed to its DNA targets by forming a ribonucleoprotein complex with these 2 small non-coding RNAs: crRNA and tracrRNA. By elegant engineering, (4zt9[7]) that too efficiently directs Cas9 protein to DNA targets encoded within the guide sequence of sgRNA [8]:
Examples of 3D structures of single guide RNA (sgRNA)
The , termed the guide sequence, adjacent to a [8][9]. Despite this, a [8][10][11][12], more so within the 5’ proximal position of the guide sequence.
- (5f9r)[15].
- .
- - place of accurate, precise, and programmable DNA cleavage.
- (sgRNA is not shown).
In the type II-A system, the Cas9-tracrRNA complex and Csn2 are involved in spacer acquisition along with the Cas1-Cas2 complex [16][17]; the involvement of Cas9 in adaptation is likely to be a general feature of type II systems. Although the key residues of Cas9 involved in PAM recognition are dispensable for spacer acquisition, they are essential for the incorporation of new spacers with the correct PAM sequence [17]. The involvement of Cas9 in PAM recognition and protospacer selection [17] suggests that in type II systems Cas1 may have lost this role.
Cas9 nuclease can be converted into (PDB entry 4zt9), an RNA-programmable DNA-binding protein, by (Figure 1b) [7][1][8][9].
In the simplest case, dCas9 can repress transcription by sterically interfering with transcription initiation or elongationby being targeted to the gene of interest with a properly chosen sgRNA [8][9][10][11][18][19][20][21][22]. The repression strength is strongly dependent on the position with respect to the target promoter as well as the nature of promoter itself [10][11][18][19]. In prokaryotes, repression of up to 1000-fold was achieved when targeting dCas9 to either DNA strand within a promoter or to the non-template DNA strand downstream. However, in eukaryotic cells such steric repression is weaker: only up to 2-fold and 20-fold repression was observed with natural promoters in mammalian and yeast cells correspondingly[10][20][21]. As a notable exception, synthetic promoters specifically constructed for direct repression by dCas9 can be repressed up to 100-fold in mammalian cells[22].
Cas9-sgRNA-target DNA complexes from Streptococcus pyogenes:
Other representatives: 5y36, 4un3.
Crystal Structure of Staphylococcus aureus Cas9[23]
The RNA-guided DNA endonuclease Cas9 cleaves double-stranded DNA targets with a protospacer adjacent motif (PAM) and complementarity to the guide RNA. Recently, we harnessed Staphylococcus aureus Cas9 (SaCas9), which is significantly smaller than Streptococcus pyogenes Cas9 (SpCas9), to facilitate efficient in vivo genome editing. Here, the crystal structures of SaCas9 in complex with a single guide RNA (sgRNA) and its double stranded DNA targets, containing the 5'-TTGAAT-3' PAM and the 5'-TTGGGT-3' PAM, at 2.6 and 2.7 A˚ resolutions, respectively, were reported. The structures revealed the mechanism of the relaxed recognition of the 5'-NNGRRT-3' PAM by SaCas9. A structural comparison of SaCas9 with SpCas9 highlighted both structural conservation and divergence, explaining their distinct PAM specificities and orthologous sgRNA recognition.
Overall Structure of the SaCas9–sgRNA–Target DNA Complex
The (residues 1–1053; N580A/C946A) in complex with a 73-nucleotide (nt) sgRNA, a 28-nt target DNA strand and an 8-nt non-target DNA strand, containing the 5'-TTGAAT-3' PAM was solved (5czz). SaCas9 adopts a consisting of a REC lobe (residues 41–425) and a NUC lobe (residues 1–40 and 435–1053). The by an arginine-rich bridge helix (residues 41–73) and a linker loop (residues 426–434). The the RuvC (residues 1–40, 435–480 and 650–774), HNH (residues 520–628), WED (residues 788–909), and PI (residues 910–1053) domains. The can be divided into a Topoisomerase-homology (TOPO) domain and a C-terminal domain (CTD). The three separate motifs (RuvC-I–III) and interacts with the HNH and PI domains. The is connected to RuvC-II and RuvC-III by the L1 (residues 481–519) and L2 (residues 629–649) linker regions, respectively. The active site of the HNH domain is distant from the cleavage site in the target DNA strand (the phosphodiester linkage between dC3 and dA4), indicating that the present structure represents the inactive state. The WED and RuvC domains are connected by a loop (residues 775–787). Previous structural studies revealed that SpCas9 undergoes conformational rearrangements upon guide RNA binding, to form the central channel between the REC and NUC lobes. In the absence of the guide RNA, SpCas9 and AnCas9 adopt a closed conformation, where the active site of the HNH domain is covered by the RuvC domain. In contrast, the ternary and quaternary complex structures of SpCas9 adopt an open conformation and have the central channel, which accommodates the guide RNA–target DNA heteroduplex (referred to as the guide:target heteroduplex). The present , suggesting that the guide RNA-induced conformational
activation is conserved between SaCas9 and SpCas9. A structural comparison between SaCas9 and SpCas9 revealed that, although their overall architectures are similar, there are notable differences in their REC, WED, and PI domains, as described in detail below, thereby explaining the significant sequence and size differences of the two Cas9 orthologs.
Structure of the sgRNA–Target DNA Complex
The SaCas9 sgRNA consists of the guide region (G1–C20),repeat region (G21–G34), tetraloop (G35–A38), anti-repeat region (C39–C54), stem loop 1 (A56–G68), and single-stranded linker (U69–U73), with A55 connecting the anti-repeat region and stem loop 1. U73 at the 3' end is disordered in the present structure. The guide region (G1–C20) and the target DNA strand (dG1–dC20) form the , whereas the target DNA strand (dC(8)–dA(1)) and the non-target DNA strand (dT1*–dG8*) form a (referred to as the PAM duplex). The repeat (G21–G34) and anti-repeat (C39–C54) regions form a distorted duplex (referred to as the ) via 13 Watson-Crick base pairs. is formed via three Watson-Crick base pairs (G57:C67–C59:G65) and two non-canonical base pairs (A56:G68 and A60:A63). U64 does not base pair with A60 and is flipped out of the stem loop. The N1 and N6 of A63 hydrogen bond with the 2'-OH and N3 of A60, respectively. G68 stacks with G57:C67, with the G68 N2 interacting with the backbone phosphate group between A55 and A56. A55 adopts the syn conformation, and its adenine base stacks with U69. In addition, the N1 of A55 hydrogen bonds with the 2'-OH of G68, thus stabilizing the basal region of stem loop 1. An adenosine residue immediately after the repeat:anti-repeat duplex is highly conserved among CRISPR-Cas9 systems, and the equivalent adenosine in the SpCas9 sgRNA, A51, also adopts the syn conformation, suggesting that these adenosine residues play conserved key roles in connecting the repeat:anti-repeat duplex and stem loop 1.
Recognition Mechanism of the Guide:Target Heteroduplex
The formed between the REC and NUC lobes. The sugar-phosphate backbone of the PAM-distal region (A3–U6) of the sgRNA interacts with the . In SpCas9 and SaCas9, the RNA–DNA base pairing in the 8 bp PAM-proximal ‘‘seed’’ region in the guide:target heteroduplex is critical for Cas9-catalyzed DNA cleavage. Consistent with this, the phosphate backbone of the sgRNA seed region (C13–C20) is extensively recognized , as in the case of SpCas9. These structural observations explain the RNA-guided DNA targeting mechanism of SaCas9. The C-terminal region of the REC lobe interacts with the PAM-distal region of the heteroduplex, whereas the N-terminal region of the REC lobe interacts with the repeat:anti-repeat duplex and the PAM-proximal region of the heteroduplex. Notably, the C-terminal region of the REC lobe of SaCas9 shares structural similarity with those of SpCas9 (PDB: 4un3, 26% identity, rmsd of 1.9 A˚ for 177 equivalent Ca atoms) and AnCas9 (PDB: 4oge, 16% identity, rmsd of 3.2 A˚ for 167 equivalent Ca atoms). These structural findings suggested that the Cas9 orthologs recognize the PAM-distal region of the guide:target heteroduplex in a similar manner.
Recognition Mechanism of the sgRNA Scaffold
The . Consistent with our data showing that the distorted repeat:anti-repeat duplex is critical for Cas9-catalyzed DNA cleavage, the . The 2'-OH of C30 hydrogen bonds with , and the backbone phosphate groups of U31, C45, and U46 interact with , respectively. These structural observations explain the structure-dependent recognition of the repeat:anti-repeat duplex by SaCas9. Stem loop 1 is recognized by the bridge helix and the REC lobe. The phosphate backbone of interacts with the bridge helix () and the REC lobe (). The . The flipped-out , respectively. A55 is extensively recognized by the phosphate lock loop. The , respectively. . The phosphate backbone of the , and the nucleobase of on the bridge helix.
Recognition Mechanism of the 5'-NNGRRT-3' PAM
SaCas9 recognizes the 5'-NNGRRN-3' PAM, with a preference for a thymine base at the 6th position, which is distinct from the 5'-NGG-3' PAM of SpCas9. In the present structures containing either the or the , the PAM duplex is sandwiched between the WED and PI domains, and the PAM in the non-target DNA strand is read from the major groove side by the PI domain. dT1* and dT2* do not directly contact the protein. Consistent with the observed requirement for the 3rd G in the 5'-NNGRRT-3' PAM, the O6 and N7 of dG3* form bidentate hydrogen bonds with the side chain of Arg1015, which is anchored via salt bridges with Glu993 in both complexes. In the 5'-TTGAAT-3' PAM complex, the , respectively. In addition, the N6 of dA5* forms a water-mediated hydrogen bond with Asn985. Similarly, in the 5'-TTGGGT-3' PAM complex, the , respectively. The O6 of dG5* forms a water-mediated hydrogen bond with Asn985. These structural features explain the ability of SaCas9 to recognize the purine nucleotides at positions 4 and 5 in the 5'-NNGRRT-3' PAM. The O4 of dT6* hydrogen bonds with Arg991, explaining the preference of SaCas9 for the 6th T in the 5'-NNGRRT-3' PAM. Single alanine mutations of these PAM-interacting residues reduced the cleavage activity in vivo, and double mutations abolished the activity, confirming the importance of Asn985, Asn986, Arg991, Glu993, and Arg1015 for PAM recognition. In addition, the phosphate backbone of the PAM duplex is recognized from the minor groove side by the WED domain (Tyr789, Tyr882, Lys886, Ans888, Ala889, and Leu909), in a distinct manner from that in SpCas9. Together, these structural and functional data have revealed the mechanism underlying the relaxed recognition of the 5'-NNGRRT-3' PAM by SaCas9.
Structural Basis for the Distinct PAM Specificities
A structural comparison of SaCas9, SpCas9, and AnCas9 revealed that, despite the lack of sequence homology, their PI domains share a similar protein fold. The PI domains consist of the TOPO domain, comprising a three-stranded anti-parallel β-sheet (β1–β3) flanked by several α-helices, and the C-terminal domain, comprising a twisted six-stranded anti-parallel β-sheet (β4–β9) (β7 in SpCas9 adopts a loop conformation). In both SaCas9 and SpCas9, the major groove of the PAM duplex is read by the β5–β7 region in their PI domains. The 3rd G in the 5'-NNGRRT-3' PAM is recognized by Arg1015 in SaCas9 , whereas the 3rd G in the 5'-NGG-3' PAM is recognized by Arg1335 in SpCas9 in a similar manner. However, there are notable differences in the PI domains of SaCas9 and SpCas9, consistent with their distinct PAM specificities. Arg1333 of SpCas9, which recognizes the 2nd G in the 5'-NGG-3' PAM, is replaced with Pro1013 in SaCas9. In addition, SpCas9 lacks the amino acid residues equivalent to Asn985/Asn986 (β5) and Arg991 (β6) of SaCas9, because the b5–b6 region of SpCas9 is shorter than that of SaCas9. Moreover, Asn985, Asn986, Arg991, and Arg1015 in SaCas9 are replaced with Asp1030, Thr1031, Lys1034, and Lys1061 in AnCas9, respectively, suggesting that the PAM of AnCas9 is different from those of SaCas9 and SpCas9 (although the sequence remains unknown). Together, these structural findings demonstrate that the distinct PAM specificities of the Cas9 orthologs are primarily defined by the specific differences in the PAM-interacting residues in the PI domains.
Mechanism of Target DNA Unwinding
In SpCas9, Glu1108 and Ser1109, in the phosphate lock loop, hydrogen bond with the phosphate group between dA(1) and dT1 in the target DNA strand (referred to as the +1 phosphate), thereby contributing to the target DNA unwinding. The present structure revealed that SaCas9 also has the phosphate lock loop, although it shares limited sequence similarity to that of SpCas9. In SaCas9, the +1 phosphate between dA(1) and dG1, in the target DNA strand, hydrogen bonds with the main-chain amide groups of Asp786 and Thr787 and the side chain of Thr787 in the phosphate lock loop. These interactions result in the rotation of the +1 phosphate, thereby facilitating base-pairing between dG1 in the target DNA strand and C20 in the sgRNA. Indeed, the SaCas9 T787A mutant showed reduced DNA cleavage activity (Figure 5C), confirming the functional significance of Thr787 in the phosphate lock loop. These observations indicated the conserved molecular mechanism of target DNA unwinding in SaCas9 and SpCas9.
RuvC and HNH Nuclease Domains
The RuvC domain of SaCas9 has an RNase H fold, and shares structural similarity with those of SpCas9 (PDB: 4un3, 26% identity, rmsd of 2.0 A˚ for 179 equivalent Ca atoms) and AnCas9 (PDB: 4oge, 17% identity, rmsd of 3.0 A˚ for 169 equivalent Ca atoms). Asp10, Glu477, His701, and Asp704 ofSaCas9 are located at positions similar to those of the catalytic
residues of SpCas9 (Asp10, Glu762, His983, and Asp986) and AnCas9 (Asp17, Glu505, His736, and Asp739). Indeed, the D10A, E477A, H701A, and D704A mutants of SaCas9 exhibited almost no DNA cleavage activity, suggesting that the SaCas9 RuvC domain
cleaves the non-target DNA strand through a two-metal ion mechanism, as in other RNase H superfamily endonucleases. The HNH domain of SaCas9 has a ββα-metal fold, and shares structural similarity with those of SpCas9 (27% identity, rmsd of 1.8 A˚ for 93 equivalent Ca atoms) and AnCas9 (18% identity, rmsd of 2.6 A˚ for 98 equivalent Ca atoms). Asp556, His557, and Asn580 of SaCas9 are located at positions similar to those of the catalytic residues of SpCas9 (Asp839, His840, and Asn863) and AnCas9 (Asp581, His582, and Asn606). Indeed, the H557A and N580A mutants of SaCas9 almost completely lacked DNA cleavage activity, suggesting that the SaCas9 HNH domain cleaves the target DNA strand through a one-metal ion mechanism, as in other ββα-metal endonucleases. A structural comparison of SaCas9 with SpCas9 and AnCas9 revealed that the RuvC and HNH domains are connected by α-helical linkers, L1 and L2, and that notable differences exist in the relative arrangements between the two nuclease domains. A biochemical study suggested that PAM duplex binding to SpCas9 facilitates the cleavage of the target DNA strand by the HNH domain. However, in the PAM-containing quaternary complex structures of SaCas9 and SpCas9, the HNH domains are distant from the cleavage site of the target DNA strand. A structural comparison of SaCas9 with Thermus thermophilus RuvC in complex with a Holliday junction substrate indicated steric clashes between the L1 linker and the modeled non-target DNA strand, bound to the active site of the SaCas9 RuvC domain. These observations suggested that the binding of the non-target DNA strand to the RuvC domain may facilitate a conformational change of L1, thereby bringing the HNH domain to the scissile phosphate group in the target DNA strand.
Structure-Guided Engineering of SaCas9 Transcription Activators and Inducible Nucleases
Using the crystal structure of SaCas9, we sought to conduct structure-guided engineering to further expand the CRISPR-Cas9
toolbox, as we have done previously using the SpCas9 crystal structure. Given the similarities in the overall domain organizations of SaCas9 and SpCas9, we initially explored the feasibility of engineering the SaCas9 sgRNA, to develop robust transcription activators. In the SpCas9 structure, the tetraloop and stem loop 2 of the sgRNA are exposed to the solvent, and permitted the insertion of RNA aptamers into the sgRNA to create robust RNA-guided transcription activators. To generate the SaCas9-based activator system, a catalytically inactive version of SaCas9 (dSaCas9) was created by introducing the D10A and N580A mutations to inactivate the RuvC and HNH domains, respectively, and attached VP64 to the C terminus of dSaCas9. The sgRNA scaffold was modified by the insertion of the MS2 aptamer stem loop (MS2-SL), to allow the recruitment of MS2-p65-HSF1 transcriptional activation modules. To evaluate the dSaCas9-based activator design, a transcriptional activation reporter system was constructed, consisting of tandem sgRNA target sites upstream of a minimal CMV promoter driving the expression of the fluorescent reporter gene mKate2. An additional transcriptional termination signal upstream of the reporter cassette was included, to reduce the background previously observed in a similar reporter. Robust activation of mKate2 transcription was observed when the engineered sgRNA complementary to the target sites was expressed, whereas the non-binding sgRNA had no detectable effect. Based on a screening of different sgRNA designs with this reporter assay, was found that the insertions of MS2-SL into the tetraloop and putative stem loop 2 induced strong activation in our reporter system, whereas the insertion of MS2-SL into stem loop 1 yielded modest activation, consistent with the structural data. The single insertion of MS2-SL into the tetraloop was the simplest design that yielded strong transcriptional activation. Using this optimal sgRNA design, we further tested the activation of endogenous targets in the human genome. Two guides were selected each for the human ASCL1 and MYOD1 genomic loci, and demonstrated that the dSaCas9-based activator system activated both genes to levels comparable to those of the dSpCas9-based activator. Given that the sgRNAs for SaCas9 and SpCas9 are not interchangeable, the SaCas9-based transcription activator platform complements the SpCas9-based activator systems, by allowing the independent activation of different sets of genes. The SpCas9 structure also facilitated the rational design of split-Cas9s, which can be further engineered into an inducible system. This SaCas9 structure revealed several flexible regions in SaCas9 that could likewise serve as potential split sites. Three versions of a split-SaCas9 were created, and two of them showed robust cleavage activity at the endogenous EMX1 target locus. Using the best split design, inducible schemes were then tested based on the abscisic acid (ABA) sensing system, as well as two versions of the rapamycin-inducible FKBP/FRB system. All three systems were able to support inducible SaCas9 cleavage activity, demonstrating the possibility of an inducible, split-SaCas9 design; however, further optimization is required to increase its efficiency and reduce its background activity.
See aslo

