We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
2vab
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
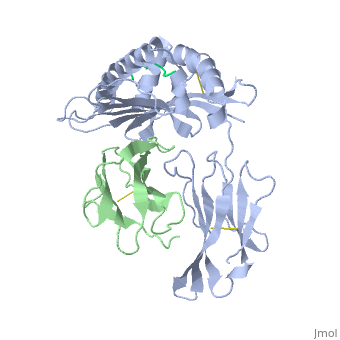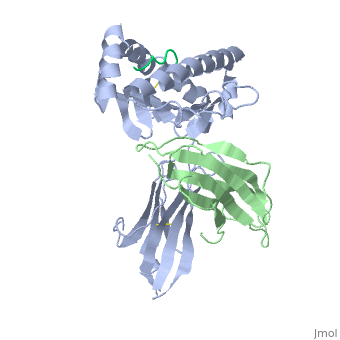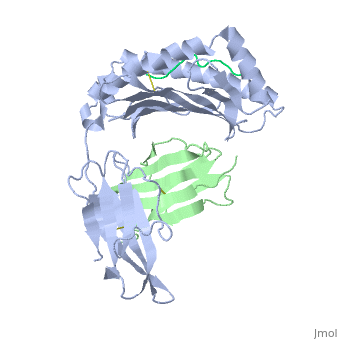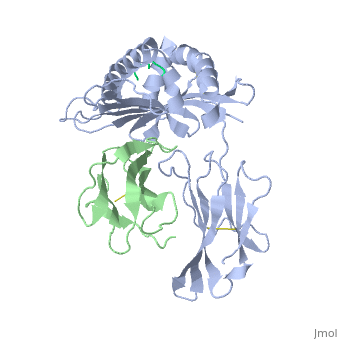
==MHC CLASS I H-2KB HEAVY CHAIN COMPLEXED WITH BETA-2 MICROGLOBULIN AND SENDAI VIRUS NUCLEOPROTEIN== | ==MHC CLASS I H-2KB HEAVY CHAIN COMPLEXED WITH BETA-2 MICROGLOBULIN AND SENDAI VIRUS NUCLEOPROTEIN== | ||
| - | <StructureSection load='2vab' size='340' side='right' caption='[[2vab]], [[Resolution|resolution]] 2.50Å' scene=''> | + | <StructureSection load='2vab' size='340' side='right'caption='[[2vab]], [[Resolution|resolution]] 2.50Å' scene=''> |
== Structural highlights == | == Structural highlights == | ||
| - | <table><tr><td colspan='2'>[[2vab]] is a 3 chain structure with sequence from [ | + | <table><tr><td colspan='2'>[[2vab]] is a 3 chain structure with sequence from [https://en.wikipedia.org/wiki/Lk3_transgenic_mice Lk3 transgenic mice] and [https://en.wikipedia.org/wiki/Murine_respirovirus Murine respirovirus]. This structure supersedes the now removed PDB entry [http://oca.weizmann.ac.il/oca-bin/send-pdb?obs=1&id=1vab 1vab]. The February 2005 RCSB PDB [https://pdb.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/index.html Molecule of the Month] feature on ''Major Histocompatibility Complex'' by David S. Goodsell is [https://dx.doi.org/10.2210/rcsb_pdb/mom_2005_2 10.2210/rcsb_pdb/mom_2005_2]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2VAB OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=2VAB FirstGlance]. <br> |
| - | </td></tr><tr id='gene'><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">BETA-2-MICROGLOBULIN ([ | + | </td></tr><tr id='gene'><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">BETA-2-MICROGLOBULIN ([https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=10090 LK3 transgenic mice])</td></tr> |
| - | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=2vab FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2vab OCA], [https://pdbe.org/2vab PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=2vab RCSB], [https://www.ebi.ac.uk/pdbsum/2vab PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=2vab ProSAT]</span></td></tr> |
</table> | </table> | ||
== Function == | == Function == | ||
| - | [[ | + | [[https://www.uniprot.org/uniprot/HA1B_MOUSE HA1B_MOUSE]] Involved in the presentation of foreign antigens to the immune system. [[https://www.uniprot.org/uniprot/NCAP_SENDE NCAP_SENDE]] Encapsidates the genome in a ratio of one N per six ribonucleotides, protecting it from nucleases. The nucleocapsid (NC) has a helical structure with 13.07 N per turn. The encapsidated genomic RNA is termed the NC and serves as template for transcription and replication. Replication is dependent on intracellular concentration of newly synthesized N, termed N(0), which corresponds to the protein not associated with RNA. In contrast, when associated with RNA, it is termed N. During replication, encapsidation by N(0) is coupled to RNA synthesis and all replicative products are resistant to nucleases. [[https://www.uniprot.org/uniprot/B2MG_MOUSE B2MG_MOUSE]] Component of the class I major histocompatibility complex (MHC). Involved in the presentation of peptide antigens to the immune system. |
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 30: | Line 30: | ||
==See Also== | ==See Also== | ||
| - | *[[Beta-2 microglobulin|Beta-2 microglobulin]] | + | *[[Beta-2 microglobulin 3D structures|Beta-2 microglobulin 3D structures]] |
*[[Cation-pi interactions|Cation-pi interactions]] | *[[Cation-pi interactions|Cation-pi interactions]] | ||
| - | *[[ | + | *[[MHC 3D structures|MHC 3D structures]] |
| - | + | ||
== References == | == References == | ||
<references/> | <references/> | ||
__TOC__ | __TOC__ | ||
</StructureSection> | </StructureSection> | ||
| + | [[Category: Large Structures]] | ||
[[Category: Lk3 transgenic mice]] | [[Category: Lk3 transgenic mice]] | ||
[[Category: Major Histocompatibility Complex]] | [[Category: Major Histocompatibility Complex]] | ||
Revision as of 10:43, 7 July 2021
MHC CLASS I H-2KB HEAVY CHAIN COMPLEXED WITH BETA-2 MICROGLOBULIN AND SENDAI VIRUS NUCLEOPROTEIN
| |||||||||||