Rhomboid protease
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
== Structural highlights == | == Structural highlights == | ||
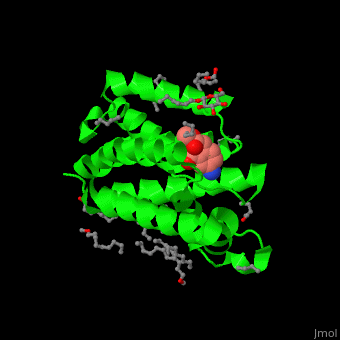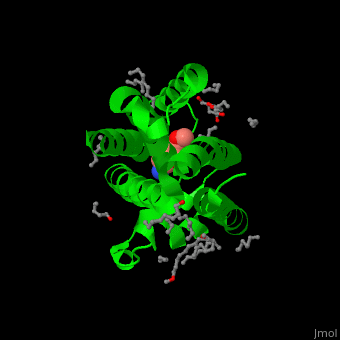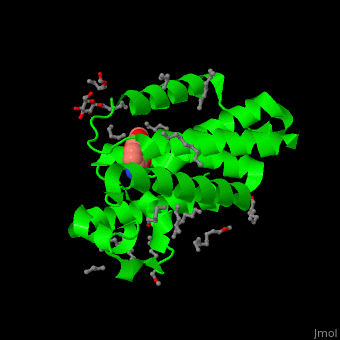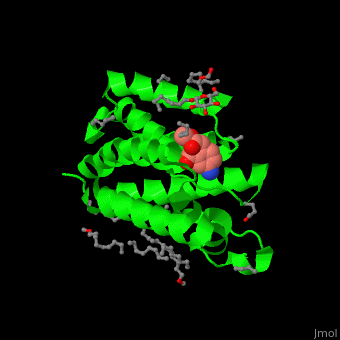
| - | The inhibitor <scene name='55/551211/Cv/ | + | The inhibitor <scene name='55/551211/Cv/5'>isocoumarin covalently binds to the GlpG catalytic residues Ser and His</scene> (in deepskyblue)<ref>PMID:20890268</ref>. <scene name='55/551211/Cv/6'>Whole active site</scene>. Water molecules shown as red spheres. |
</StructureSection> | </StructureSection> | ||
Revision as of 13:18, 27 August 2019
| |||||||||||
3D Structures of rhomboid protease
Updated on 27-August-2019
References
- ↑ Freeman M. Rhomboid proteases and their biological functions. Annu Rev Genet. 2008;42:191-210. doi: 10.1146/annurev.genet.42.110807.091628. PMID:18605900 doi:http://dx.doi.org/10.1146/annurev.genet.42.110807.091628
- ↑ Vinothkumar KR, Strisovsky K, Andreeva A, Christova Y, Verhelst S, Freeman M. The structural basis for catalysis and substrate specificity of a rhomboid protease. EMBO J. 2010 Oct 1. PMID:20890268 doi:10.1038/emboj.2010.243